Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes
- PMID: 14595104
- PMCID: PMC329406
- DOI: 10.1091/mbc.e03-08-0594
Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes
Abstract
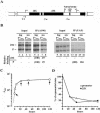
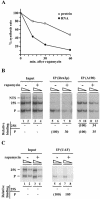
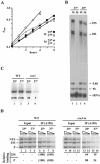
Yeast cells entering into stationary phase decrease rRNA synthesis rate by decreasing both the number of active genes and the transcription rate of individual active genes. Using chromatin immunoprecipitation assays, we found that the association of RNA polymerase I with the promoter and the coding region of rDNA is decreased in stationary phase, but association of transcription factor UAF with the promoter is unchanged. Similar changes were also observed when growing cells were treated with rapamycin, which is known to inhibit the Tor signaling system. Rapamycin treatment also caused a decrease in the amount of Rrn3p-polymerase I complex, similar to stationary phase. Because recruitment of Pol I to the rDNA promoter is Rrn3p-dependent as shown in this work, these data suggest that the decrease in the transcription rate of individual active genes in stationary phase is achieved by the Tor signaling system acting at the Rrn3p-dependent polymerase recruitment step. Miller chromatin spreads of cells treated with rapamycin and cells in post-log phase confirm this conclusion and demonstrate that the Tor system does not participate in alteration of the number of active genes observed for cells entering into stationary phase.
Figures
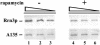


References
-
- Broach, J.R. (1991). RAS genes in Saccharomyces cerevisiae: signal transduction in search of a pathway. Trends Genet. 7, 28-33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

