Cytoscape: a software environment for integrated models of biomolecular interaction networks
- PMID: 14597658
- PMCID: PMC403769
- DOI: 10.1101/gr.1239303
Cytoscape: a software environment for integrated models of biomolecular interaction networks
Abstract
Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework. Although applicable to any system of molecular components and interactions, Cytoscape is most powerful when used in conjunction with large databases of protein-protein, protein-DNA, and genetic interactions that are increasingly available for humans and model organisms. Cytoscape's software Core provides basic functionality to layout and query the network; to visually integrate the network with expression profiles, phenotypes, and other molecular states; and to link the network to databases of functional annotations. The Core is extensible through a straightforward plug-in architecture, allowing rapid development of additional computational analyses and features. Several case studies of Cytoscape plug-ins are surveyed, including a search for interaction pathways correlating with changes in gene expression, a study of protein complexes involved in cellular recovery to DNA damage, inference of a combined physical/functional interaction network for Halobacterium, and an interface to detailed stochastic/kinetic gene regulatory models.
Figures


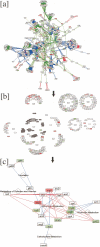

References
-
- Batagelj, V. and Mrvar, A. 1998. Pajek—Program for large network analysis. Connections 21: 47-57.
-
- Begley, T.J., Rosenbach, A.S., Ideker, T., and Samson, L.D. 2002. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 1: 103-112. - PubMed
-
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680-686. - PubMed
WEB SITE REFERENCES
-
- http://biodata.mshri.on.ca/; Osprey Network Visualization System
-
- http://pim.hybrigenics.com/; PIMRider
-
- http://predictome.bu.edu/; Predictome Project
-
- http://www.cytoscape.org/; Cytoscape v1.1 Home Page
-
- http://www.cytoscape.org/plugins/SBW/; Supplementary data on model exchange via SBML
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
