Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment
- PMID: 14597712
- PMCID: PMC263858
- DOI: 10.1073/pnas.1735529100
Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment
Abstract
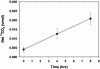
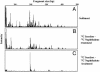
Microorganisms maintain the biosphere by catalyzing biogeochemical processes, including biodegradation of organic chemical pollutants. Yet seldom have the responsible agents and their respective genes been identified. Here we used field-based stable isotopic probing (SIP) to discover a group of bacteria responsible for in situ metabolism of an environmental pollutant, naphthalene. We released 13C-labeled naphthalene in a contaminated study site to trace the flow of pollutant carbon into the naturally occurring microbial community. Using GC/MS, molecular biology, and classical microbiological techniques we documented 13CO2 evolution (2.3% of the dose in 8 h), created a library of 16S rRNA gene clones from 13C labeled sediment DNA, identified a taxonomic cluster (92 of 95 clones) from the microbial community involved in metabolism of the added naphthalene, and isolated a previously undescribed bacterium (strain CJ2) from site sediment whose 16S rRNA gene matched that of the dominant member (48%) of the clone library. Strain CJ2 is a beta proteobacterium closely related to Polaromonas vacuolata. Moreover, strain CJ2 hosts the sequence of a naphthalene dioxygenase gene, prevalent in site sediment, detected before only in environmental DNA. This investigative strategy may have general application for elucidating the bases of many biogeochemical processes, hence for advancing knowledge and management of ecological and industrial systems that rely on microorganisms.
Figures



Similar articles
-
Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site.Appl Environ Microbiol. 1997 Jun;63(6):2330-7. doi: 10.1128/aem.63.6.2330-2337.1997. Appl Environ Microbiol. 1997. PMID: 9172352 Free PMC article.
-
Horizontal transfer of phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments.Appl Environ Microbiol. 2003 Apr;69(4):2172-81. doi: 10.1128/AEM.69.4.2172-2181.2003. Appl Environ Microbiol. 2003. PMID: 12676698 Free PMC article.
-
Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA.Appl Environ Microbiol. 2003 Mar;69(3):1614-22. doi: 10.1128/AEM.69.3.1614-1622.2003. Appl Environ Microbiol. 2003. PMID: 12620850 Free PMC article.
-
The role of active-site residues in naphthalene dioxygenase.J Ind Microbiol Biotechnol. 2003 May;30(5):271-8. doi: 10.1007/s10295-003-0043-3. Epub 2003 Apr 15. J Ind Microbiol Biotechnol. 2003. PMID: 12695887 Review.
-
Microbial analysis of soil and groundwater from a gasworks site and comparison with a sequenced biological reactive barrier remediation process.J Appl Microbiol. 2007 May;102(5):1227-38. doi: 10.1111/j.1365-2672.2007.03398.x. J Appl Microbiol. 2007. PMID: 17448158 Review.
Cited by
-
Draft Genome Sequence of a Metabolically Diverse Antarctic Supraglacial Stream Organism, Polaromonas sp. Strain CG9_12, Determined Using Pacific Biosciences Single-Molecule Real-Time Sequencing Technology.Genome Announc. 2014 Dec 4;2(6):e01242-14. doi: 10.1128/genomeA.01242-14. Genome Announc. 2014. PMID: 25477404 Free PMC article.
-
Characterization of novel polycyclic aromatic hydrocarbon dioxygenases from the bacterial metagenomic DNA of a contaminated soil.Appl Environ Microbiol. 2014 Nov;80(21):6591-600. doi: 10.1128/AEM.01883-14. Epub 2014 Aug 15. Appl Environ Microbiol. 2014. PMID: 25128340 Free PMC article.
-
Benzene Degradation by a Variovorax Species within a Coal Tar-Contaminated Groundwater Microbial Community.Appl Environ Microbiol. 2017 Feb 1;83(4):e02658-16. doi: 10.1128/AEM.02658-16. Print 2017 Feb 15. Appl Environ Microbiol. 2017. PMID: 27913419 Free PMC article.
-
Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy.Front Microbiol. 2015 Mar 31;6:242. doi: 10.3389/fmicb.2015.00242. eCollection 2015. Front Microbiol. 2015. PMID: 25873913 Free PMC article. Review.
-
Construction and Evaluation of a Korean Native Microbial Consortium for the Bioremediation of Diesel Fuel-Contaminated Soil in Korea.Front Microbiol. 2018 Oct 30;9:2594. doi: 10.3389/fmicb.2018.02594. eCollection 2018. Front Microbiol. 2018. PMID: 30425703 Free PMC article.
References
-
- Radajewski, S., Ineson, R., Parekh, N. R. & Murrell, J. C. (2000) Nature 403, 646-649. - PubMed
-
- Beja, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C., Feldman, R. A. & DeLong, E. F. (2000) Science 289, 1902-1906. - PubMed
-
- Newman, D. K. & Banfield, J. F. (2002) Science 296, 1071-1077. - PubMed
-
- Jetten, M. S. M, Strous, M., van de Pas-Schoonen, K. T., Schalk, J., van Dongen, J. G. J. M., Van de Graaf, A. A., Logemaann, S., Muyzer, G., van Loosedrecht, M. C. M. & Kuenen, J. G. (1998) FEMS Microbiol. Rev. 22, 421-437. - PubMed
-
- Madsen, E. L. (1998) Environ. Sci. Technol. 32, 429-439.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

