Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome
- PMID: 14597773
- PMCID: PMC2173662
- DOI: 10.1083/jcb.200304040
Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome
Abstract
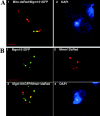
The unit of inheritance for mitochondrial DNA (mtDNA) is a complex nucleoprotein structure termed the nucleoid. The organization of the nucleoid as well as its role in mtDNA replication remain largely unknown. Here, we show in Saccharomyces cerevisiae that at least two populations of nucleoids exist within the same mitochondrion and can be distinguished by their association with a discrete proteinaceous structure that spans the outer and inner mitochondrial membranes. Surprisingly, this two membrane-spanning structure (TMS) persists and self-replicates in the absence of mtDNA. We tested whether TMS functions to direct the replication of mtDNA. By monitoring BrdU incorporation, we observed that actively replicating nucleoids are associated exclusively with TMS. Consistent with TMS's role in mtDNA replication, we found that Mip1, the mtDNA polymerase, is also a stable component of TMS. Taken together, our observations reveal the existence of an autonomous two membrane-spanning mitochondrial replisome as well as provide a mechanism for how mtDNA replication and inheritance may be physically linked.
Figures



Similar articles
-
The TFAM-to-mtDNA ratio defines inner-cellular nucleoid populations with distinct activity levels.Cell Rep. 2021 Nov 23;37(8):110000. doi: 10.1016/j.celrep.2021.110000. Cell Rep. 2021. PMID: 34818548
-
Yeast model analysis of novel polymerase gamma variants found in patients with autosomal recessive mitochondrial disease.Hum Genet. 2015 Sep;134(9):951-66. doi: 10.1007/s00439-015-1578-x. Epub 2015 Jun 16. Hum Genet. 2015. PMID: 26077851 Free PMC article.
-
A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery.Mol Biol Cell. 2003 Nov;14(11):4618-27. doi: 10.1091/mbc.e03-04-0225. Epub 2003 Sep 17. Mol Biol Cell. 2003. PMID: 13679517 Free PMC article.
-
Organization of DNA in Mammalian Mitochondria.Int J Mol Sci. 2019 Jun 5;20(11):2770. doi: 10.3390/ijms20112770. Int J Mol Sci. 2019. PMID: 31195723 Free PMC article. Review.
-
To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins.Biochimie. 2014 May;100:219-26. doi: 10.1016/j.biochi.2013.09.017. Epub 2013 Sep 25. Biochimie. 2014. PMID: 24076128 Review.
Cited by
-
Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast.J Cell Biol. 2005 Jan 3;168(1):103-15. doi: 10.1083/jcb.200410030. J Cell Biol. 2005. PMID: 15631992 Free PMC article.
-
Making heads or tails of phospholipids in mitochondria.J Cell Biol. 2011 Jan 10;192(1):7-16. doi: 10.1083/jcb.201006159. J Cell Biol. 2011. PMID: 21220505 Free PMC article. Review.
-
Contribution of ERMES subunits to mature peroxisome abundance.PLoS One. 2019 Mar 25;14(3):e0214287. doi: 10.1371/journal.pone.0214287. eCollection 2019. PLoS One. 2019. PMID: 30908556 Free PMC article.
-
Shaping the dynamic mitochondrial network.BMC Biol. 2014 May 27;12:35. doi: 10.1186/1741-7007-12-35. BMC Biol. 2014. PMID: 24884775 Free PMC article. Review.
-
Eukaryotic DNA Polymerases in Homologous Recombination.Annu Rev Genet. 2016 Nov 23;50:393-421. doi: 10.1146/annurev-genet-120215-035243. Annu Rev Genet. 2016. PMID: 27893960 Free PMC article. Review.
References
-
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie, III, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. - PubMed
-
- Bevis, B.J., and B.S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83–87. (published erratum appears in Nat. Biotechnol. 2002. 20:1159) - PubMed
-
- Birky, C.W., Jr. 1978. Transmission genetics of mitochondria and choroplasts. Annu. Rev. Genet. 12:471–512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

