Cold shock response and major cold shock proteins of Vibrio cholerae
- PMID: 14602587
- PMCID: PMC262262
- DOI: 10.1128/AEM.69.11.6361-6369.2003
Cold shock response and major cold shock proteins of Vibrio cholerae
Abstract
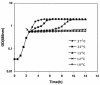
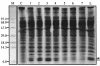
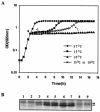
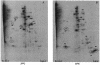
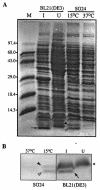
When exponentially growing Vibrio cholerae cells were shifted from 37 degrees C to various lower temperatures, it was found that the organism could adapt and grow at temperatures down to 15 degrees C, below which the growth was completely arrested. There was no difference between the patterns of the cold shock responses in toxinogenic and nontoxinogenic strains of V. cholerae. Gel electrophoretic analyses of proteins of cold-exposed cells revealed significant induction of two major cold shock proteins (Csps), whose molecular masses were 7.7 kDa (CspA(VC)) and 7.5 kDa (CspV), and six other Csps, most of which were much larger. We cloned, sequenced, and analyzed the cspV gene encoding the CspV protein of V. cholerae O139 strain SG24. Although CspA(VC) and CspV have similar kinetics of synthesis and down-regulation, the corresponding genes, cspA and cspV, which are located in the small chromosome, are not located in the same operon. A comparative analysis of the kinetics of synthesis revealed that the CspV protein was synthesized de novo only during cold shock. Although both CspA(VC) and CspV were stable for several hours in the cold, the CspV protein was degraded rapidly when the culture was shifted back to 37 degrees C, suggesting that this protein is probably necessary for adaptation at lower temperatures. Northern blot analysis confirmed that the cspV gene is cold shock inducible and is regulated tightly at the level of transcription. Interestingly, the cspV gene has a cold shock-inducible promoter which is only 12 nucleotides from the translational start site, and therefore, it appears that no unusually long 5' untranslated region is present in its mRNA transcript. Thus, this promoter is an exception compared to other promoters of cold shock-inducible genes of different organisms, including Escherichia coli. Our results suggest that V. cholerae may use an alternative pathway for regulation of gene expression during cold shock.
Figures



References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
-
- Bhadra, R. K., S. Roychoudhury, and J. Das. 1994. Vibrio cholerae O139 biotype El Tor. Lancet 343:728. - PubMed
-
- Carroll, W. J., M. C. Mateescu, K. Chava, R. R. Colwell, and A. K. Bej. 2001. Response and tolerance of toxigenic Vibrio cholerae O1 to cold temperatures. Antonie Leeuwenhoek 79:377-384. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

