Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences
- PMID: 14602635
- PMCID: PMC262318
- DOI: 10.1128/AEM.69.11.6740-6749.2003
Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences
Abstract
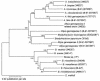
Bacteria belonging to the genera Afipia and Bosea are amoeba-resisting bacteria that have been recently reported to colonize hospital water supplies and are suspected of being responsible for intensive care unit-acquired pneumonia. Identification of these bacteria is now based on determination of the 16S ribosomal DNA sequence. However, the 16S rRNA gene is not polymorphic enough to ensure discrimination of species defined by DNA-DNA relatedness. The complete rpoB sequences of 20 strains were first determined by both PCR and genome walking methods. The percentage of homology between different species ranged from 83 to 97% and was in all cases lower than that observed with the 16S rRNA gene; this was true even for species that differed in only one position. The taxonomy of Bosea and Afipia is discussed in light of these results. For strain identification that does not require the complete rpoB sequence (4,113 to 4,137 bp), we propose a simple computerized method that allows determination of nucleotide positions of high variability in the sequence that are bordered by conserved sequences and that could be useful for design of universal primers. A fragment of 740 to 752 bp that contained the most highly variable area (positions 408 to 420) was amplified and sequenced with these universal primers for 47 strains. The variability of this sequence allowed identification of all strains and correlated well with results of DNA-DNA relatedness. In the future, this method could be also used for the determination of variability "hot spots" in sets of housekeeping genes, not only for identification purposes but also for increasing the discriminatory power of sequence typing techniques such as multilocus sequence typing.
Figures


References
-
- Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. - PubMed
-
- Barker, J., and M. R. W. Brown. 1994. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 140:1253-1259. - PubMed
-
- Brenner, D. J., D. G. Hollis, C. W. Moss, C. K. English, G. S. Hall, J. Vincent, J. Radosevic, K. A. Birkness, W. F. Bibb, F. D. Quinn, B. Swaminathan, R. E. Weaver, M. W. Reeves, S. P. O'Connor, P. S. Hayes, F. C. Tenover, A. G. Steigerwalt, B. A. Perkins, M. L. Daneshvar, B. C. Hill, J. A. Washington, T. C. Woods, S. B. Hunter, T. D. Hadfield, G. W. Ajello, A. F. Kaufmann, D. J. Wear, and J. D. Wenger. 1991. Proposal of Afipia gen. nov. with Afipia felis gen. nov. sp. nov. (formerly the cat scratch bacillus), Afipia clevelandensis sp. nov. (formerly the Cleveland clinic foundation strain), Afipia broomeae sp. nov., and three unnamed genospecies. J. Clin. Microbiol. 29:2450-2460. - PMC - PubMed
-
- Das, S. K., A. K. Mishra, B. J. Tindall, F. A. Rainey, and E. Stackebrandt. 1996. Oxidation of thiosulfate by a new bacterium, Bosea thiooxidans (strain BI-42) gen. nov., sp. nov.: analysis of phylogeny based on chemotaxonomy and 16S ribosomal DNA sequencing. Int. J. Syst. Bacteriol. 46:981-987. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

