In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp)
- PMID: 14602911
- PMCID: PMC275556
- DOI: 10.1093/nar/gkg860
In vitro RNP assembly and methylation guide activity of an unusual box C/D RNA, cis-acting archaeal pre-tRNA(Trp)
Abstract
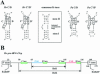
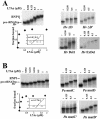
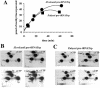
Among the large family of C/D methylation guide RNAs, the intron of euryarchaeal pre-tRNA(Trp) represents an outstanding specimen able to guide in cis, instead of in trans, two 2'-O-methylations in the pre-tRNA exons. Remarkably, both sites of methylation involve nucleotides within the bulge-helix-bulge (BHB) splicing motif, while the RNA-guided methylation and pre-tRNA splicing events depend on mutually exclusive RNA folding patterns. Using the three recombinant core proteins of archaeal C/D RNPs, we have analyzed in vitro RNP assembly of the pre-tRNA and tested its site-specific methylation activity. Recognition by L7Ae of hallmark K-turns at the C/D and C'/D' motifs appears as a crucial assembly step required for subsequent binding of a Nop5p-aFib heterodimer at each site. Unexpectedly, however, even without L7Ae but at a higher concentration of Nop5p-aFib, a substantially active RNP complex can still form, possibly reflecting the higher propensity of the cis-acting system to form guide RNA duplex(es) relative to classical trans- acting C/D RNA guides. Moreover, footprinting data of RNPs, consistent with Nop5p interacting with the non-canonical stem of the K-turn, suggest that binding of Nop5p-aFib to the pre-tRNA-L7Ae complex might direct transition from a splicing-competent structure to an RNA conformer displaying the guide RNA duplexes required for site-specific methylation.
Figures



Similar articles
-
Sequential 2'-O-methylation of archaeal pre-tRNATrp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA.J Biol Chem. 2004 Nov 12;279(46):47661-71. doi: 10.1074/jbc.M408868200. Epub 2004 Sep 3. J Biol Chem. 2004. PMID: 15347671
-
Dynamic guide-target interactions contribute to sequential 2'-O-methylation by a unique archaeal dual guide box C/D sRNP.RNA. 2008 Jul;14(7):1411-23. doi: 10.1261/rna.1003308. Epub 2008 May 30. RNA. 2008. PMID: 18515549 Free PMC article.
-
Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp.Nucleic Acids Res. 2001 Nov 15;29(22):4518-29. doi: 10.1093/nar/29.22.4518. Nucleic Acids Res. 2001. PMID: 11713301 Free PMC article.
-
Noncoding RNAs of the H/ACA family.Cold Spring Harb Symp Quant Biol. 2006;71:395-405. doi: 10.1101/sqb.2006.71.034. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381322 Review.
-
Small non-coding RNAs in Archaea.Curr Opin Microbiol. 2005 Dec;8(6):685-94. doi: 10.1016/j.mib.2005.10.013. Epub 2005 Oct 26. Curr Opin Microbiol. 2005. PMID: 16256421 Review.
Cited by
-
Euryarchaeal beta-CASP proteins with homology to bacterial RNase J Have 5'- to 3'-exoribonuclease activity.J Biol Chem. 2010 Jun 4;285(23):17574-83. doi: 10.1074/jbc.M109.095117. Epub 2010 Apr 7. J Biol Chem. 2010. PMID: 20375016 Free PMC article.
-
Dissecting the role of conserved box C/D sRNA sequences in di-sRNP assembly and function.Nucleic Acids Res. 2010 Dec;38(22):8295-305. doi: 10.1093/nar/gkq690. Epub 2010 Aug 6. Nucleic Acids Res. 2010. PMID: 20693534 Free PMC article.
-
RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes.BMC Genomics. 2008 Oct 9;9:470. doi: 10.1186/1471-2164-9-470. BMC Genomics. 2008. PMID: 18844986 Free PMC article.
-
Induced fit of RNA on binding the L7Ae protein to the kink-turn motif.RNA. 2005 Aug;11(8):1192-200. doi: 10.1261/rna.2680605. Epub 2005 Jun 29. RNA. 2005. PMID: 15987806 Free PMC article.
-
Transfer RNA Modification Enzymes from Thermophiles and Their Modified Nucleosides in tRNA.Microorganisms. 2018 Oct 20;6(4):110. doi: 10.3390/microorganisms6040110. Microorganisms. 2018. PMID: 30347855 Free PMC article. Review.
References
-
- Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 255–272.
-
- Bachellerie J.P., Cavaille,J. and Huttenhofer,A. (2002) The expanding snoRNA world. Biochimie, 84, 775–790. - PubMed
-
- Dennis P.P., Omer,A. and Lowe,T. (2001) A guided tour: small RNA function in Archaea. Mol. Microbiol., 40, 509–519. - PubMed
-
- Filipowicz W. and Pogacic,V. (2002) Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol., 14, 319–327. - PubMed

