Enzyme-specific profiles for genome annotation: PRIAM
- PMID: 14602924
- PMCID: PMC275543
- DOI: 10.1093/nar/gkg847
Enzyme-specific profiles for genome annotation: PRIAM
Abstract
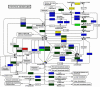
The advent of fully sequenced genomes opens the ground for the reconstruction of metabolic pathways on the basis of the identification of enzyme-coding genes. Here we describe PRIAM, a method for automated enzyme detection in a fully sequenced genome, based on the classification of enzymes in the ENZYME database. PRIAM relies on sets of position-specific scoring matrices ('profiles') automatically tailored for each ENZYME entry. Automatically generated logical rules define which of these profiles is required in order to infer the presence of the corresponding enzyme in an organism. As an example, PRIAM was applied to identify potential metabolic pathways from the complete genome of the nitrogen-fixing bacterium Sinorhizobium meliloti. The results of this automated method were compared with the original genome annotation and visualised on KEGG graphs in order to facilitate the interpretation of metabolic pathways and to highlight potentially missing enzymes.
Figures

References
-
- Bork P., Ouzounis,C., Casari,G., Schneider,R., Sander,C., Dolan,M., Gilbert,W. and Gillevet,P.M. (1995) Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol. Microbiol., 16, 955–967. - PubMed
-
- Tatusov R.L., Mushegian,A.R., Bork,P., Brown,N.P., Hayes,W.S., Borodovsky,M., Rudd,K.E. and Koonin,E.V. (1996) Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr. Biol., 6, 279–291. - PubMed
-
- Fitch W.M. (2000) Homology a personal view on some of the problems. Trends Genet., 16, 227–231. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

