Yeast life-span extension by calorie restriction is independent of NAD fluctuation
- PMID: 14605207
- PMCID: PMC4998187
- DOI: 10.1126/science.1088697
Yeast life-span extension by calorie restriction is independent of NAD fluctuation
Abstract
Calorie restriction (CR) slows aging in numerous species. In the yeast Saccharomyces cerevisiae, this effect requires Sir2, a conserved NAD+-dependent deacetylase. We report that CR reduces nuclear NAD+ levels in vivo. Moreover, the activity of Sir2 and its human homologue SIRT1 are not affected by physiological alterations in the NAD+:NADH ratio. These data implicate alternate mechanisms of Sir2 regulation by CR.
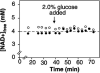
Figures


References
-
- Lin SJ, Defossez PA, Guarente L. Science. 2000;289:2126. - PubMed
-
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. FASEB J. 2000;14:2135. - PubMed
-
- Swiecilo A, Krawiec Z, Wawryn J, Bartosz G, Bilinski T. Acta Biochim. Polym. 2000;47:355. - PubMed
-
- Smith JS, et al. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6658. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

