Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells
- PMID: 14608036
- PMCID: PMC299905
- DOI: 10.1073/pnas.2334901100
Mouse glucocorticoid-induced tumor necrosis factor receptor ligand is costimulatory for T cells
Abstract
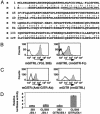
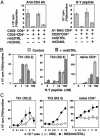
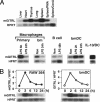
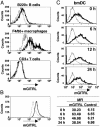
Recently, agonist antibodies to glucocorticoid-induced tumor necrosis factor receptor (GITR) (tumor necrosis factor receptor superfamily 18) have been shown to neutralize the suppressive activity of CD4+CD25+ regulatory T cells. It was anticipated that this would be the role of the physiological ligand. We have identified and expressed the gene for mouse GITR ligand and have confirmed that its interaction with GITR reverses suppression by CD4+CD25+ T cells. It also, however, provides a costimulatory signal for the antigen-driven proliferation of naïve T cells and polarized T helper 1 and T helper 2 clones. RT-PCR and mAb staining revealed mouse GITR ligand expression in dendritic cells, macrophages, and B cells. Expression was controlled by the transcription factor NF-1 and potentially by alternative splicing of mRNA destabilization sequences.
Figures

Comment in
-
Costimulatory signals controlling regulatory T cells.Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15292-3. doi: 10.1073/pnas.0307001100. Epub 2003 Dec 15. Proc Natl Acad Sci U S A. 2003. PMID: 14676329 Free PMC article. No abstract available.
References
-
- Gurney, A. L., Marsters, S. A., Huang, R. M., Pitti, R. M., Mark, D. T., Baldwin, D. T., Gray, A. M., Dowd, A. D., Brush, A. D., Heldens, A. D., et al. (1999) Curr. Biol. 9, 215-218. - PubMed
-
- Kwon, B., Yu, K.-Y., Ni, J., Yu, G.-L., Jang, I.-K., Kim, Y.-J., Xing, L., Liu, D., Wang, S.-X. & Kwon, B. S. (1999) J. Biol. Chem. 274, 6056-6061. - PubMed
-
- McHugh, R. S., Whitters, M. J., Piccirillo, C. A., Young, D. A., Shevach, E. M., Collins, M. & Byrne, M. C. (2002) Immunity 16, 311-323. - PubMed
-
- Shimizu, J., Yamazaki, S., Takahashi, T., Ishida, Y. & Sakaguchi, S. (2002) Nat. Immunol. 3, 135-142. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

