LIME, a novel transmembrane adaptor protein, associates with p56lck and mediates T cell activation
- PMID: 14610044
- PMCID: PMC2194117
- DOI: 10.1084/jem.20030232
LIME, a novel transmembrane adaptor protein, associates with p56lck and mediates T cell activation
Abstract
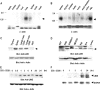
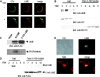
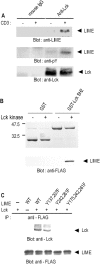
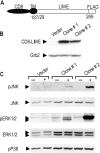
In this study, we identify and characterize a novel transmembrane adaptor protein, designated Lck-interacting membrane protein (LIME), as a binding partner of the Lck Src homology (SH)2 domain. LIME possesses a short extracellular domain, a transmembrane domain, and a cytoplasmic tail containing five tyrosine-based motifs. The protein is primarily expressed in hematopoietic cells and lung. Interestingly, LIME expression is up-regulated by TCR stimulation and sustained up to 24 h, suggesting that LIME acts throughout the early to late stages of T cell activation. LIME is localized to membrane rafts and distributed within the T cell-APC contact site. Upon TCR stimulation of Jurkat T cells, LIME associates with Lck as a tyrosine-phosphorylated protein. Experiments using Jurkat T cells expressing CD8-LIME chimera reveal that the protein associates with phosphatidylinositol 3-kinase, Grb2, Gads, and SHP2, and activates ERK1/2 and JNK but not p38. Moreover, overexpression of LIME in Jurkat T cells induces transcriptional activation of the IL-2 promoter. Our data collectively show that LIME is a raft-associated transmembrane adaptor protein linking TCR stimuli to downstream signaling pathways via associations with Lck.
Figures


References
-
- Wange, R.L., and L.E. Samelson. 1996. Complex complexes: signaling at the TCR. Immunity. 5:197–205. - PubMed
-
- Weiss, A., and D.R. Littman. 1994. Signal transduction by lymphocyte antigen receptors. Cell. 76:263–274. - PubMed
-
- Cherukuri, A., M. Dykstra, and S.K. Pierce. 2001. Floating the raft hypothesis: lipid rafts play a role in immune cell activation. Immunity. 14(6):657–660. - PubMed
-
- Zhang, W., J. Sloan-Lancaster, J. Kitchen, R.P. Trible, and L.E. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 92:83–92. - PubMed
-
- Wange R.L. 2000. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci. STKE. 2000:RE1. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

