Relationship between RNA lariat debranching and Ty1 element retrotransposition
- PMID: 14610201
- PMCID: PMC262579
- DOI: 10.1128/jvi.77.23.12795-12806.2003
Relationship between RNA lariat debranching and Ty1 element retrotransposition
Abstract
The Saccharomyces cerevisiae DBR1 gene encodes a 2'-5' phosphodiesterase that debranches intron RNA lariats following splicing. Yeast dbr1 mutants accumulate intron lariats and are also defective for mobility of the retrotransposons Ty1 and Ty3. We used a mutagenic PCR method to generate a collection of dbr1 mutant alleles to explore the relationship between the roles of DBR1 in transposition and debranching. Eight mutants defective for Ty1 transposition contained single amino acid changes in Dbr1p. Two mutations, G84A and N85D, are in a conserved phosphoesterase motif that is believed to be part of the active site of the enzyme, supporting a connection between enzymatic activity and Ty1 transposition. Two other mutations, Y68F and Y68D, occur at a potential phosphorylation site, and we have shown that Dbr1p is phosphorylated on tyrosine. We have developed an RNase protection assay to quantitate intron RNA accumulation in cells. The assay uses RNA probes that hybridize to ACT1 intron RNA. Protection patterns confirm that sequences from the 5' end of the intron to the lariat branch point accumulate in dbr1 mutants in a branched (lariat) conformation. RNase protection assays indicate that all of the newly generated dbr1 mutant alleles are also deficient for debranching, further supporting a role for 2'-5' phosphodiesterase activity in Ty1 transposition. A Ty1 element lacking most of its internal sequences transposes independently of DBR1. The existence of Dbr1p-dependent Ty1 sequences raises the possibility that Dbr1p acts on Ty1 RNA.
Figures





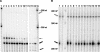

References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2003. Current protocols in molecular biology on CD-ROM. Current Protocols Inc., Brooklyn, N.Y.
-
- Boeke, J., and J. Stoye. 1997. Retrotransposons, endogenous retroviruses and the evolution of retroelements, p. 343-435. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. - PubMed
-
- Boeke, J. D., F. Lacroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. - PubMed
-
- Cadwell, R. C., and G. F. Joyce. 1994. Mutagenic PCR. PCR Methods Appl. 3:S136-S140. - PubMed
-
- Cadwell, R. C., and G. F. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28-33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

