Receptor-activated binding of viral fusion proteins to target membranes
- PMID: 14610829
- PMCID: PMC7119201
- DOI: 10.1016/S0076-6879(03)72026-6
Receptor-activated binding of viral fusion proteins to target membranes
Abstract
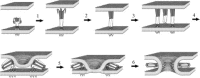
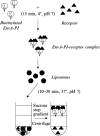
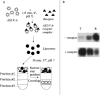
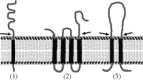
This chapter describes three assays to monitor receptor-induced association of the envelope glycoprotein (EnvA) of avian sarcoma/leukosis virus (ASLV) with target bilayers: (1) the original assay for monitoring binding of the EnvA ectodomain (EnvA-PI) to target membranes (liposomes), (2) a modified and miniaturized EnvA-PI-liposome binding assay, and (3) an assay to measure binding of intact sarcoma/leukosis virus subtype A (ASLV-A) virus particles to target membranes. These assays are also useful for studying other receptor-activated viral fusion proteins. When one viral glycoprotein and one “simple” host cell receptor are involved, it should be possible to develop assays directly analogous to those described above for studying Tva-induced binding of the EnvA ectodomain (EnvA-PI) to target membranes. A general prerequisite for a fusion protein/target membrane binding assay is a soluble and correctly oligomeric form of the viral fusion protein ectodomain. The simplest host cell receptors that would be amenable to this type of analysis are type I or type II integral membrane proteins. The soluble versions of the ectodomains of these receptors, produced by genetic engineering or proteolytic release, could then be used to trigger the cognate fusion protein. The methodology could, similarly, be applicable to multimembrane-spanning host cell receptors when the functional part of the receptor is tethered at only one end or where an ectodomain loop preserves enough structure to function as a soluble analog, perhaps by generating a cyclic peptide analog of the loop. The same “receptor reagents” could be employed for intact virus particle/target membrane binding assays.
Figures

Similar articles
-
Liposomes as target membranes in the study of virus receptor interaction and membrane fusion.Methods Enzymol. 2003;372:374-92. doi: 10.1016/S0076-6879(03)72022-9. Methods Enzymol. 2003. PMID: 14610825 No abstract available.
-
Virus membrane-fusion proteins: more than one way to make a hairpin.Nat Rev Microbiol. 2006 Jan;4(1):67-76. doi: 10.1038/nrmicro1326. Nat Rev Microbiol. 2006. PMID: 16357862 Free PMC article. Review.
-
Pore formation in target liposomes by viral fusion proteins.Methods Enzymol. 2003;372:408-18. doi: 10.1016/S0076-6879(03)72024-2. Methods Enzymol. 2003. PMID: 14610827 No abstract available.
-
Liposome fusion assay to monitor intracellular membrane fusion machines.Methods Enzymol. 2003;372:274-300. doi: 10.1016/S0076-6879(03)72016-3. Methods Enzymol. 2003. PMID: 14610819 No abstract available.
-
Virus membrane fusion.FEBS Lett. 2007 May 22;581(11):2150-5. doi: 10.1016/j.febslet.2007.01.093. Epub 2007 Feb 16. FEBS Lett. 2007. PMID: 17320081 Free PMC article. Review.
Cited by
-
Receptor-induced thiolate couples Env activation to retrovirus fusion and infection.PLoS Pathog. 2007 Dec 21;3(12):e198. doi: 10.1371/journal.ppat.0030198. PLoS Pathog. 2007. PMID: 18260686 Free PMC article.
-
Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion.Proc Natl Acad Sci U S A. 2007 Nov 20;104(47):18718-23. doi: 10.1073/pnas.0707452104. Epub 2007 Nov 14. Proc Natl Acad Sci U S A. 2007. PMID: 18003913 Free PMC article.
-
Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane.Mol Biol Cell. 2005 Dec;16(12):5502-13. doi: 10.1091/mbc.e05-06-0496. Epub 2005 Sep 29. Mol Biol Cell. 2005. PMID: 16195349 Free PMC article.
-
Stable association of herpes simplex virus with target membranes is triggered by low pH in the presence of the gD receptor, HVEM.J Virol. 2006 Apr;80(8):3773-80. doi: 10.1128/JVI.80.8.3773-3780.2006. J Virol. 2006. PMID: 16571794 Free PMC article.
-
The avian retrovirus avian sarcoma/leukosis virus subtype A reaches the lipid mixing stage of fusion at neutral pH.J Virol. 2003 Mar;77(5):3058-66. doi: 10.1128/jvi.77.5.3058-3066.2003. J Virol. 2003. PMID: 12584331 Free PMC article.
References
-
- Bullough P.A, Hughson F.M, Skehel J.J, Wiley D.C. Nature. 1994;371:37. - PubMed
-
- Rey F.A, Heinz F.X, Mandl C, Kunz C, Harrison S.C. Nature. 1995;375:291. - PubMed
-
- Lescar J, Roussel A, Wien M.W, Navaza J, Fuller S.D, Wengler G, Rey F.A. Cell. 2001;105:137. - PubMed
-
- Heinz F.X, Allison S.L. Curr. Opin. Microbiol. 2001;4:450. - PubMed
-
- Carr C.M, Kim P.S. Cell. 1993;73:823. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

