Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts
- PMID: 14612416
- PMCID: PMC262674
- DOI: 10.1128/MCB.23.23.8762-8772.2003
Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts
Abstract
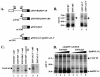
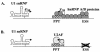
Human immunodeficiency virus type 1 (HIV-1) exonic splicing silencers (ESSs) inhibit production of certain spliced viral RNAs by repressing alternative splicing of the viral precursor RNA. Several HIV-1 ESSs interfere with spliceosome assembly by binding cellular hnRNP A/B proteins. Here, we have further characterized the mechanism of splicing repression using a representative HIV-1 hnRNP A/B-dependent ESS, ESSV, which regulates splicing at the vpr 3' splice site. We show that hnRNP A/B proteins bound to ESSV are necessary to inhibit E complex assembly by competing with the binding of U2AF65 to the polypyrimidine tracts of repressed 3' splice sites. We further show evidence suggesting that U1 snRNP binds the 5' splice site despite an almost complete block of splicing by ESSV. Possible splicing-independent functions of U1 snRNP-5' splice site interactions during virus replication are discussed.
Figures

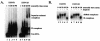

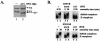

Similar articles
-
RNA splicing at human immunodeficiency virus type 1 3' splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element.J Virol. 2001 Sep;75(18):8487-97. doi: 10.1128/jvi.75.18.8487-8497.2001. J Virol. 2001. PMID: 11507194 Free PMC article.
-
The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element.EMBO J. 2001 Oct 15;20(20):5748-58. doi: 10.1093/emboj/20.20.5748. EMBO J. 2001. PMID: 11598017 Free PMC article.
-
hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure.RNA. 2002 Nov;8(11):1401-15. doi: 10.1017/s1355838202023075. RNA. 2002. PMID: 12458794 Free PMC article.
-
Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing.Curr HIV Res. 2006 Jan;4(1):43-55. doi: 10.2174/157016206775197655. Curr HIV Res. 2006. PMID: 16454710 Review.
-
How Are Short Exons Flanked by Long Introns Defined and Committed to Splicing?Trends Genet. 2016 Oct;32(10):596-606. doi: 10.1016/j.tig.2016.07.003. Epub 2016 Aug 6. Trends Genet. 2016. PMID: 27507607 Review.
Cited by
-
Let It Go: HIV-1 cis-Acting Repressive Sequences.J Virol. 2021 Jul 12;95(15):e0034221. doi: 10.1128/JVI.00342-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 33980600 Free PMC article. Review.
-
Evidence for cooperative tandem binding of hnRNP C RRMs in mRNA processing.RNA. 2015 Nov;21(11):1931-42. doi: 10.1261/rna.052373.115. Epub 2015 Sep 14. RNA. 2015. PMID: 26370582 Free PMC article.
-
An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA.J Virol. 2013 Mar;87(5):2707-20. doi: 10.1128/JVI.02755-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255806 Free PMC article.
-
Tra2-mediated recognition of HIV-1 5' splice site D3 as a key factor in the processing of vpr mRNA.J Virol. 2013 Mar;87(5):2721-34. doi: 10.1128/JVI.02756-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255807 Free PMC article.
-
Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5' end.Nucleic Acids Res. 2005 Feb 8;33(3):825-37. doi: 10.1093/nar/gki185. Print 2005. Nucleic Acids Res. 2005. PMID: 15701754 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
