The DIVa maturase binding site in the yeast group II intron aI2 is essential for intron homing but not for in vivo splicing
- PMID: 14612420
- PMCID: PMC262681
- DOI: 10.1128/MCB.23.23.8809-8819.2003
The DIVa maturase binding site in the yeast group II intron aI2 is essential for intron homing but not for in vivo splicing
Abstract
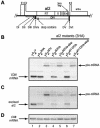
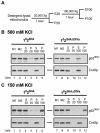
Splicing of the Saccharomyces cerevisiae mitochondrial DNA group II intron aI2 depends on the intron-encoded 62-kDa reverse transcriptase-maturase protein (p62). In wild-type strains, p62 remains associated with the excised intron lariat RNA in ribonucleoprotein (RNP) particles that are essential for intron homing. Studies of a bacterial group II intron showed that the DIVa substructure of intron domain IV is a high-affinity binding site for its maturase. Here we first present in vitro evidence extending that conclusion to aI2. Then, experiments with aI2 DIVa mutant strains show that the binding of p62 to DIVa is not essential for aI2 splicing in vivo but is essential for homing. Because aI2 splicing in the DIVa mutant strains remains maturase dependent, splicing must rely on other RNA-protein contacts. The p62 that accumulates in the mutant strains has reverse transcriptase activity, but fractionation experiments at high and low salt concentrations show that it associates more weakly than the wild-type protein with endogenous mitochondrial RNAs, and that phenotype probably explains the homing defect. Replacing the DIVa of aI2 with that of the closely related intron aI1 improves in vivo splicing but not homing, indicating that DIVa contributes to the specificity of the maturase-RNA interaction needed for homing.
Figures




References
-
- Arnberg, A., G. V. Ommen, L. Grivell, E. V. Bruggen, and P. Borst. 1980. Some yeast mitochondrial RNAs are circular. Cell 19:313-319. - PubMed
-
- Belfort, M., V. Derbyshire, M. M. Parker, B. Cousineau, and A. M. Lambowitz. 2002. Mobile introns: pathways and proteins, p. 761-783. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
-
- Bonitz, S. G., G. Coruzzi, B. E. Thalenfield, A. Tzagoloff, and G. Macino. 1980. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit I of cytochrome oxidase. J. Biol. Chem. 255:11927-11941. - PubMed
-
- Boudvillain, M., A. de Lencastre, and A. M. Pyle. 2000. A tertiary interaction that links active-site domains to the 5′ splice site of a group II intron. Nature 406:315-318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
