Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences
- PMID: 14612421
- PMCID: PMC262689
- DOI: 10.1128/MCB.23.23.8820-8828.2003
Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences
Abstract
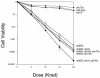
End joining of double-strand breaks (DSBs) requires Ku proteins and frequently involves base pairing between complementary terminal sequences. To define the role of terminal base pairing in end joining, two oppositely oriented HO endonuclease cleavage sites separated by 2.0 kb were integrated into yeast chromosome III, where constitutive expression of HO endonuclease creates two simultaneous DSBs with no complementary end sequence. Lack of complementary sequence in their 3' single-strand overhangs facilitates efficient repair events distinctly different from when the 3' ends have a 4-bp sequence base paired in various ways to create 2- to 3-bp insertions. Repair of noncomplementary ends results in a set of nonrandom deletions of up to 302 bp, annealed by imperfect microhomology of about 8 to 10 bp at the junctions. This microhomology-mediated end joining (MMEJ) is Ku independent, but strongly dependent on Mre11, Rad50, and Rad1 proteins and partially dependent on Dnl4 protein. The MMEJ also occurs when Rad52 is absent, but the extent of deletions becomes more limited. The increased gamma ray sensitivity of rad1Delta rad52Delta yku70Delta strains compared to rad52Delta yku70Delta strains suggests that MMEJ also contributes to the repair of DSBs induced by ionizing radiation.
Figures


References
-
- Chen, L., K. Trujillo, W. Ramos, P. Sung, and A. E. Tomkinson. 2001. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol. Cell 8:1105-1115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
