Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break
- PMID: 14612428
- PMCID: PMC262690
- DOI: 10.1128/MCB.23.23.8913-8923.2003
Yeast Rad52 and Rad51 recombination proteins define a second pathway of DNA damage assessment in response to a single double-strand break
Abstract
Saccharomyces cells with a single unrepaired double-strand break adapt after checkpoint-mediated G(2)/M arrest. We have found that both Rad51 and Rad52 recombination proteins play key roles in adaptation. Cells lacking Rad51p fail to adapt, but deleting RAD52 suppresses rad51Delta. rad52Delta also suppresses adaptation defects of srs2Delta mutants but not those of yku70Delta or tid1Delta mutants. Neither rad54Delta nor rad55Delta affects adaptation. A Rad51 mutant that fails to interact with Rad52p is adaptation defective; conversely, a C-terminal truncation mutant of Rad52p, impaired in interaction with Rad51p, is also adaptation defective. In contrast, rad51-K191A, a mutation that abolishes recombination and results in a protein that does not bind to single-stranded DNA (ssDNA), supports adaptation, as do Rad51 mutants impaired in interaction with Rad54p or Rad55p. An rfa1-t11 mutation in the ssDNA binding complex RPA partially restores adaptation in rad51Delta mutants and fully restores adaptation in yku70Delta and tid1Delta mutants. Surprisingly, although neither rfa1-t11 nor rad52Delta mutants are adaptation defective, the rad52Delta rfa1-t11 double mutant fails to adapt and exhibits the persistent hyperphosphorylation of the DNA damage checkpoint protein Rad53 after HO induction. We suggest that monitoring of the extent of DNA damage depends on independent binding of RPA and Rad52p to ssDNA, with Rad52p's activity modulated by Rad51p whereas RPA's action depends on Tid1p.
Figures
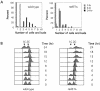
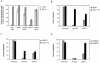




References
-
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. - PubMed
-
- Bai, Y., and L. S. Symington. 1996. A RAD52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 10:2025-2037. - PubMed
-
- de la Torre-Ruiz, M., and N. F. Lowndes. 2000. The Saccharomyces cerevisiae DNA damage checkpoint is required for efficient repair of double strand breaks by non-homologous end joining. FEBS Lett. 467:311-315. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
