RNA backbone is rotameric
- PMID: 14612579
- PMCID: PMC283519
- DOI: 10.1073/pnas.1835769100
RNA backbone is rotameric
Abstract
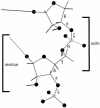
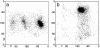
Despite the importance of local structural detail to a mechanistic understanding of RNA catalysis and binding functions, RNA backbone conformation has been quite recalcitrant to analysis. There are too many variable torsion angles per residue, and their raw empirical distributions are poorly clustered. This study applies quality-filtering techniques (using resolution, crystallographic B factor, and all-atom steric clashes) to the backbone torsion angle distributions from an 8,636-residue RNA database. With noise levels greatly reduced, clear signal appears for the underlying angle preferences. Half-residue torsion angle distributions for alpha-beta-gamma and for delta-epsilon-zeta are plotted and contoured in 3D; each shows about a dozen distinct peaks, which can then be combined in pairs to define complete RNA backbone conformers. Traditional nucleic acid residues are defined from phosphate to phosphate, but here we use a base-to-base (or sugar-to-sugar) division into "suites" to parse the RNA backbone repeats, both because most backbone steric clashes are within suites and because the relationship of successive bases is both reliably determined and conformationally important. A suite conformer has seven variables, with sugar pucker specified at both ends. Potential suite conformers were omitted if not represented by at least a small cluster of convincing data points after application of quality filters. The final result is a small library of 42 RNA backbone conformers, which should provide valid conformations for nearly all RNA backbone encountered in experimental structures.
Figures



References
-
- Kruger, K., Grabowski, P. J., Zaug, A. J., Sands, J., Gottschling, D. E. & Cech, T. R. (1982) Cell 31, 147–157. - PubMed
-
- Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. (2000) Science 289, 920–930. - PubMed
-
- Joyce, G. F. (2002) Nature 418, 214–221. - PubMed
-
- Davis, P. W., Adamiak, R. W. & Tinoco, I. J. (1990) Biopolymers 29, 109–122. - PubMed
-
- Frank, J. (2001) BioEssays 23, 725–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

