Genome-wide characterization of the lignification toolbox in Arabidopsis
- PMID: 14612585
- PMCID: PMC523881
- DOI: 10.1104/pp.103.026484
Genome-wide characterization of the lignification toolbox in Arabidopsis
Abstract
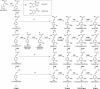
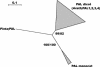
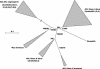
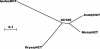
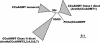
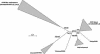
Lignin, one of the most abundant terrestrial biopolymers, is indispensable for plant structure and defense. With the availability of the full genome sequence, large collections of insertion mutants, and functional genomics tools, Arabidopsis constitutes an excellent model system to profoundly unravel the monolignol biosynthetic pathway. In a genome-wide bioinformatics survey of the Arabidopsis genome, 34 candidate genes were annotated that encode genes homologous to the 10 presently known enzymes of the monolignol biosynthesis pathway, nine of which have not been described before. By combining evolutionary analysis of these 10 gene families with in silico promoter analysis and expression data (from a reverse transcription-polymerase chain reaction analysis on an extensive tissue panel, mining of expressed sequence tags from publicly available resources, and assembling expression data from literature), 12 genes could be pinpointed as the most likely candidates for a role in vascular lignification. Furthermore, a possible novel link was detected between the presence of the AC regulatory promoter element and the biosynthesis of G lignin during vascular development. Together, these data describe the full complement of monolignol biosynthesis genes in Arabidopsis, provide a unified nomenclature, and serve as a basis for further functional studies.
Figures



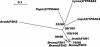


References
-
- Achnine L, Rasmussen S, Blancaflor E, Dixon RA (2002) Metabolic channeling at the entry point into the phenylpropanoid pathway: physical association between l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase. In I El Hadrami, ed, Proceedings of the XXI International Conference on Polyphenols, Imprimerie El-Watania, Marrakech, Morocco, pp 7–8
-
- AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 - PubMed
-
- Anterola AM, Jeon J-H, Davin LB, Lewis NG (2002) Transcriptional control of monolignol biosynthesis in Pinus taeda: factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J Biol Chem 277: 18272–18280 - PubMed
-
- Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

