Efficient target-selected mutagenesis in zebrafish
- PMID: 14613981
- PMCID: PMC403812
- DOI: 10.1101/gr.1725103
Efficient target-selected mutagenesis in zebrafish
Abstract
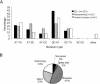
One of the most powerful methods available to assign function to a gene is to inactivate or knockout the gene. Recently,we described the first target-selected knockout in zebrafish. Here,we report on the further improvements of this procedure,resulting in a highly efficient and easy method to do target-selected mutagenesis in zebrafish. A library of 4608 ENU-mutagenized F1 animals was generated and kept as a living stock. The DNA of these animals was screened for mutations in 16 genes by use of CEL-I-mediated heteroduplex cleavage (TILLING) and subsequent resequencing. In total,255 mutations were identified,of which 14 resulted in a premature stop codon,7 in a splice donor/acceptor site mutation,and 119 in an amino acid change. By this method,we potentially knocked out 13 different genes in a few months time. Furthermore,we show that TILLING can be used to detect the full spectrum of ENU-induced mutations in a vertebrate genome with the presence of many naturally occurring polymorphisms.
Figures


References
-
- Beier, D.R. 2000. Sequence-based analysis of mutagenized mice. Mamm. Genome 11: 594-597. - PubMed
-
- Berezikov, E., Plasterk, R.H., and Cuppen, E. 2002. GENOTRACE: cDNA-based local GENOme assembly from TRACE archives. Bioinformatics 18: 1396-1397. - PubMed
-
- Coghill, E.L., Hugill, A., Parkinson, N., Davison, C., Glenister, P., Clements, S., Hunter, J., Cox, R.D., and Brown, S.D. 2002. A gene-driven approach to the identification of ENU mutants in the mouse. Nat. Genet. 30: 255-256. - PubMed
WEB SITE REFERENCES
-
- http://cuppen.niob.knaw.nl; Detailed protocols for TILLING procedures and CEL-I isolation.
-
- http://genotrace.niob.knaw.nl; Interface to GENOTRACE for local genomic sequence assemblies from raw genome sequencing traces using EST or cDNA sequences as input.
-
- http://primers.niob.knaw.nl; Design of sets of nested (tailed) primers for TILLING.
-
- http://zfin.org; The zebrafish information network.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
