The dual nature of human extracellular superoxide dismutase: one sequence and two structures
- PMID: 14615576
- PMCID: PMC283514
- DOI: 10.1073/pnas.2436143100
The dual nature of human extracellular superoxide dismutase: one sequence and two structures
Abstract
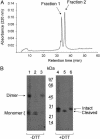
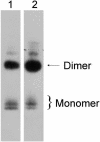
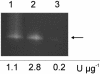
Human extracellular superoxide dismutase (EC-SOD; EC 1.15.1.1) is a scavenger of superoxide anions in the extracellular space. The amino acid sequence is homologous to the intracellular counterpart, Cu/Zn superoxide dismutase (Cu/Zn-SOD), apart from N- and C-terminal extensions. Cu/Zn-SOD is a homodimer containing four cysteine residues within each subunit, and EC-SOD is a tetramer composed of two disulfide-bonded dimers in which each subunit contains six cysteines. The amino acid sequences of all EC-SOD subunits are identical. It is known that Cys-219 is involved in an interchain disulfide. To account for the remaining five cysteine residues we purified human EC-SOD and determined the disulfide bridge pattern. The results show that human EC-SOD exists in two forms, each with a unique disulfide bridge pattern. One form (active EC-SOD) is enzymatically active and contains a disulfide bridge pattern similar to Cu/Zn-SOD. The other form (inactive EC-SOD) has a different disulfide bridge pattern and is enzymatically inactive. The EC-SOD polypeptide chain apparently folds in two different ways, most likely resulting in different three-dimensional structures. Our study shows that one gene may produce proteins with different disulfide bridge arrangements and, thus, by definition, different primary structures. This observation adds another dimension to the functional annotation of the proteome.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

