Comprehensive transposon mutant library of Pseudomonas aeruginosa
- PMID: 14617778
- PMCID: PMC283593
- DOI: 10.1073/pnas.2036282100
Comprehensive transposon mutant library of Pseudomonas aeruginosa
Abstract
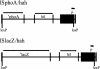
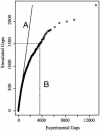
We have developed technologies for creating saturating libraries of sequence-defined transposon insertion mutants in which each strain is maintained. Phenotypic analysis of such libraries should provide a virtually complete identification of nonessential genes required for any process for which a suitable screen can be devised. The approach was applied to Pseudomonas aeruginosa, an opportunistic pathogen with a 6.3-Mbp genome. The library that was generated consists of 30,100 sequence-defined mutants, corresponding to an average of five insertions per gene. About 12% of the predicted genes of this organism lacked insertions; many of these genes are likely to be essential for growth on rich media. Based on statistical analyses and bioinformatic comparison to known essential genes in E. coli, we estimate that the actual number of essential genes is 300-400. Screening the collection for strains defective in two defined multigenic processes (twitching motility and prototrophic growth) identified mutants corresponding to nearly all genes expected from earlier studies. Thus, phenotypic analysis of the collection may produce essentially complete lists of genes required for diverse biological activities. The transposons used to generate the mutant collection have added features that should facilitate downstream studies of gene expression, protein localization, epistasis, and chromosome engineering.
Figures



References
-
- Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. - PubMed
-
- Berg, C. M. & Berg, D. E. (1995) in Mobile Genetic Elements, ed. Sherratt, D. (Oxford Univ. Press, Oxford), pp. 38-68.
-
- Kumar, A., des Etages, S. A., Coelho, P. S., Roeder, G. S. & Snyder, M. (2000) Methods Enzymol. 328, 550-574. - PubMed
-
- Ross-Macdonald, P., Coelho, P. S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K. H., Sheehan, A., Symoniatis, D., Umansky, L., et al. (1999) Nature 402, 413-418. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

