The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye
- PMID: 14617811
- PMCID: PMC329262
- DOI: 10.1091/mbc.e03-06-0374
The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye
Abstract
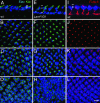
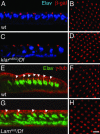
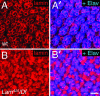
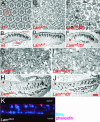
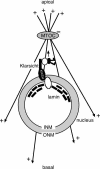
Photoreceptor nuclei in the Drosophila eye undergo developmentally regulated migrations. Nuclear migration is known to require the perinuclear protein Klarsicht, but the function of Klarsicht has been obscure. Here, we show that Klarsicht is required for connecting the microtubule organizing center (MTOC) to the nucleus. In addition, in a genetic screen for klarsicht-interacting genes, we identified Lam Dm(0), which encodes nuclear lamin. We find that, like Klarsicht, lamin is required for photoreceptor nuclear migration and for nuclear attachment to the MTOC. Moreover, perinuclear localization of Klarsicht requires lamin. We propose that nuclear migration requires linkage of the MTOC to the nucleus through an interaction between microtubules, Klarsicht, and lamin. The Klarsicht/lamin interaction provides a framework for understanding the mechanistic basis of human laminopathies.
Figures
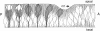


Similar articles
-
Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye.Curr Biol. 1999 Nov 4;9(21):1211-20. doi: 10.1016/s0960-9822(99)80501-6. Curr Biol. 1999. PMID: 10556085
-
Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye.Fly (Austin). 2007 Mar-Apr;1(2):75-85. doi: 10.4161/fly.4254. Fly (Austin). 2007. PMID: 18820457
-
Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye.Genetics. 2004 Nov;168(3):1385-93. doi: 10.1534/genetics.104.028662. Genetics. 2004. PMID: 15579692 Free PMC article.
-
LEM-Domain proteins: new insights into lamin-interacting proteins.Int Rev Cytol. 2007;261:1-46. doi: 10.1016/S0074-7696(07)61001-8. Int Rev Cytol. 2007. PMID: 17560279 Review.
-
Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation.Annu Rev Biochem. 2015;84:131-64. doi: 10.1146/annurev-biochem-060614-034115. Epub 2015 Feb 26. Annu Rev Biochem. 2015. PMID: 25747401 Review.
Cited by
-
Laminopathies: multiple disorders arising from defects in nuclear architecture.J Biosci. 2006 Sep;31(3):405-21. doi: 10.1007/BF02704113. J Biosci. 2006. PMID: 17006023 Review.
-
How plants LINC the SUN to KASH.Nucleus. 2013 May-Jun;4(3):206-15. doi: 10.4161/nucl.24088. Epub 2013 May 13. Nucleus. 2013. PMID: 23680964 Free PMC article. Review.
-
Understanding the roles of nuclear A- and B-type lamins in brain development.J Biol Chem. 2012 May 11;287(20):16103-10. doi: 10.1074/jbc.R112.354407. Epub 2012 Mar 13. J Biol Chem. 2012. PMID: 22416132 Free PMC article. Review.
-
Emergence of a prestressed eukaryotic nucleus during cellular differentiation and development.J R Soc Interface. 2010 Jun 6;7 Suppl 3(Suppl 3):S321-30. doi: 10.1098/rsif.2010.0039.focus. Epub 2010 Mar 31. J R Soc Interface. 2010. PMID: 20356876 Free PMC article.
-
Laminopathies: what can humans learn from fruit flies.Cell Mol Biol Lett. 2018 Jul 6;23:32. doi: 10.1186/s11658-018-0093-1. eCollection 2018. Cell Mol Biol Lett. 2018. PMID: 30002683 Free PMC article. Review.
References
-
- Apel, E.D., Lewis, R.M., Grady, R.M., and Sanes, J.R. (2000). Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986-31995. - PubMed
-
- Beckwith, S.M., Roghi, C.H., and Morris, N.R. (1995). The genetics of nuclear migration in fungi. Genet. Eng. 17, 165-180. - PubMed
-
- Bloom, K. (2000). It's a kar9ochore to capture microtubules. Nat. Cell Biol. 6, E96-E98. - PubMed
-
- Bloom, K. (2001). Nuclear migration: cortical anchors for cytoplasmic dynein. Curr. Biol. 11, R326-R329. - PubMed
-
- Burke, B., and Stewart, C.L. (2002). Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell. Biol. 3, 575-585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

