La, PTB, and PAB proteins bind to the 3(') untranslated region of Norwalk virus genomic RNA
- PMID: 14623338
- PMCID: PMC7111188
- DOI: 10.1016/j.bbrc.2003.10.066
La, PTB, and PAB proteins bind to the 3(') untranslated region of Norwalk virus genomic RNA
Abstract
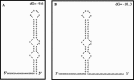
Noroviruses are human enteric caliciviruses for which no cell culture is available. Consequently, the mechanisms and factors involved in their replication have been difficult to study. In an attempt to analyze the cis- and trans-acting factors that could have a role in NV replication, the 3(')-untranslated region of the genome was studied. Use of Zuker's mfold-2 software predicted that NV 3(')UTR contains a stem-loop structure of 47 nts. Proteins from HeLa cell extracts, such as La and PTB, form stable complexes with this region. The addition of a poly(A) tail (24 nts) to the 3(')UTR permits the specific binding of the poly(A) binding protein (PABP) present in HeLa cell extracts, as well as the recombinant PABP. Since La, PTB, and PABP are important trans-acting factors required for viral translation and replication, these RNA-protein interactions may play a role in NV replication or translation.
Figures




Similar articles
-
Interaction of cellular proteins with the 5' end of Norwalk virus genomic RNA.J Virol. 2000 Sep;74(18):8558-62. doi: 10.1128/jvi.74.18.8558-8562.2000. J Virol. 2000. PMID: 10954557 Free PMC article.
-
Translation elongation factor-1alpha, La, and PTB interact with the 3' untranslated region of dengue 4 virus RNA.Virology. 2002 Apr 10;295(2):337-47. doi: 10.1006/viro.2002.1407. Virology. 2002. PMID: 12033793
-
Pyrimidine tract binding protein and La autoantigen interact differently with the 5' untranslated regions of lentiviruses and oncoretrovirus mRNAs.FEBS Lett. 2001 Feb 9;490(1-2):54-8. doi: 10.1016/s0014-5793(01)02137-8. FEBS Lett. 2001. PMID: 11172810
-
Determination of the secondary structure of and cellular protein binding to the 3'-untranslated region of the hepatitis C virus RNA genome.J Virol. 1997 Nov;71(11):8698-706. doi: 10.1128/JVI.71.11.8698-8706.1997. J Virol. 1997. PMID: 9343228 Free PMC article.
-
Functions of the 5'- and 3'-untranslated regions of tobamovirus RNA.Virus Res. 2015 Aug 3;206:82-9. doi: 10.1016/j.virusres.2015.01.028. Epub 2015 Feb 12. Virus Res. 2015. PMID: 25683511 Review.
Cited by
-
Identification of RNA-protein interaction networks involved in the norovirus life cycle.J Virol. 2012 Nov;86(22):11977-90. doi: 10.1128/JVI.00432-12. Epub 2012 Aug 29. J Virol. 2012. PMID: 22933270 Free PMC article.
-
Functional analysis of RNA structures present at the 3' extremity of the murine norovirus genome: the variable polypyrimidine tract plays a role in viral virulence.J Virol. 2010 Mar;84(6):2859-70. doi: 10.1128/JVI.02053-09. Epub 2010 Jan 6. J Virol. 2010. PMID: 20053745 Free PMC article.
-
Recent insights into reverse genetics of norovirus.Virus Res. 2023 Feb;325:199046. doi: 10.1016/j.virusres.2023.199046. Epub 2023 Jan 16. Virus Res. 2023. PMID: 36657615 Free PMC article. Review.
-
Therapeutics and Immunoprophylaxis Against Noroviruses and Rotaviruses: The Past, Present, and Future.Curr Drug Metab. 2018;19(3):170-191. doi: 10.2174/1389200218666170912161449. Curr Drug Metab. 2018. PMID: 28901254 Free PMC article. Review.
-
Cis-acting RNA elements in human and animal plus-strand RNA viruses.Biochim Biophys Acta. 2009 Sep-Oct;1789(9-10):495-517. doi: 10.1016/j.bbagrm.2009.09.007. Epub 2009 Sep 23. Biochim Biophys Acta. 2009. PMID: 19781674 Free PMC article. Review.
References
-
- Fields N.B., Knipe D.M., Howley P.M., editors. Human Caliciviruses. Lippincott, Williams, and Wilkins (Ed.); Philadelphia, USA: 2001. pp. 841–874. (Virology, fourth ed.).
-
- Frankhouser R.L., Noel J.S., Monroe S.S., Ando T., Glass R.I. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 1998;178:1571–1578. - PubMed
-
- Subekti D.S., Tjaniadi P., Lsmana M., McArdle J., Iskandriati D., Budiarsa I.N., Walujo P., Suparto H.I., Winoto I., Campbell J.R., Porter K.R., Sajuthi K.D., Ansari A.A., Oyofo B.A. Experimental infection of Macaca nemestrina with a Toronto Norwalk-like virus of epidemic viral gastroenteritis. J. Med. Virol. 2002;66:400–406. - PubMed
-
- Jiang X., Wang X.M., Wang K., Estes M.K. Sequence and genomic organization of norwalk virus. Virology. 1993;195:51–61. - PubMed
-
- Clarke N., Lambden P.R. The molecular biology of caliciviruses. J. Gen. Virol. 1997;78:291–301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

