A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease
- PMID: 14623910
- PMCID: PMC2194120
- DOI: 10.1084/jem.20031220
A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease
Abstract
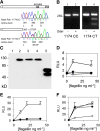
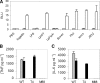
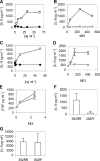
Although Toll-like receptors (TLRs) are critical mediators of the immune response to pathogens, the influence of polymorphisms in this gene family on human susceptibility to infection is poorly understood. We demonstrated recently that TLR5 recognizes flagellin, a potent inflammatory stimulus present in the flagellar structure of many bacteria. Here, we show that a common stop codon polymorphism in the ligand-binding domain of TLR5 (TLR5392STOP) is unable to mediate flagellin signaling, acts in a dominant fashion, and is associated with susceptibility to pneumonia caused by Legionella pneumophila, a flagellated bacterium. We also show that flagellin is a principal stimulant of proinflammatory cytokine production in lung epithelial cells. Together, these observations suggest that TLR5392STOP increases human susceptibility to infection through an unusual dominant mechanism that compromises TLR5's essential role as a regulator of the lung epithelial innate immune response.
Figures
References
-
- Bartlett, J.G., and L.M. Mundy. 1995. Community-acquired pneumonia. N. Engl. J. Med. 333:1618–1624. - PubMed
-
- Cooke, G.S., and A.V. Hill. 2001. Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2:967–977. - PubMed
-
- Aderem, A., and R.J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature. 406:782–787. - PubMed
-
- Janeway, C.A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197–216. - PubMed
-
- Poltorak, A., X. He, I. Smirnova, M.Y. Liu, C.V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, et al. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 282:2085–2088. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

