Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA
- PMID: 14624239
- PMCID: PMC261875
- DOI: 10.1371/journal.pbio.0000033
Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5' recessed DNA
Abstract
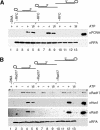
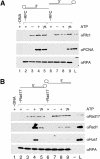
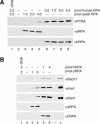
The cellular pathways involved in maintaining genome stability halt cell cycle progression in the presence of DNA damage or incomplete replication. Proteins required for this pathway include Rad17, Rad9, Hus1, Rad1, and Rfc-2, Rfc-3, Rfc-4, and Rfc-5. The heteropentamer replication factor C (RFC) loads during DNA replication the homotrimer proliferating cell nuclear antigen (PCNA) polymerase clamp onto DNA. Sequence similarities suggest the biochemical functions of an RSR (Rad17-Rfc2-Rfc3-Rfc4-Rfc5) complex and an RHR heterotrimer (Rad1-Hus1-Rad9) may be similar to that of RFC and PCNA, respectively. RSR purified from human cells loads RHR onto DNA in an ATP-, replication protein A-, and DNA structure-dependent manner. Interestingly, RSR and RFC differed in their ATPase activities and displayed distinct DNA substrate specificities. RSR preferred DNA substrates possessing 5' recessed ends whereas RFC preferred 3' recessed end DNA substrates. Characterization of the biochemical loading reaction executed by the checkpoint clamp loader RSR suggests new insights into the mechanisms underlying recognition of damage-induced DNA structures and signaling to cell cycle controls. The observation that RSR loads its clamp onto a 5' recessed end supports a potential role for RHR and RSR in diverse DNA metabolism, such as stalled DNA replication forks, recombination-linked DNA repair, and telomere maintenance, among other processes.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

