Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity
- PMID: 14627811
- PMCID: PMC290259
- DOI: 10.1093/nar/gkg891
Solution structure and DNA binding of the effector domain from the global regulator PrrA (RegA) from Rhodobacter sphaeroides: insights into DNA binding specificity
Abstract
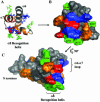
Prr/RegA response regulator is a global transcription regulator in purple bacteria Rhodobacter sphaeroides and Rhodobacter capsulatus, and is essential in controlling the metabolic changes between aerobic and anaerobic environments. We report here the structure determination by NMR of the C-terminal effector domain of PrrA, PrrAC. It forms a three-helix bundle containing a helix-turn-helix DNA binding motif. The fold is similar to FIS protein, but the domain architecture is different from previously characterised response regulator effector domains, as it is shorter than any characterised so far. Alignment of Prr/RegA DNA targets permitted a refinement of the consensus sequence, which contains two GCGNC inverted repeats with variable half-site spacings. NMR titrations of PrrAC with specific and non-specific DNA show which surfaces are involved in DNA binding and suggest residues important for binding specificity. A model of the PrrAC/DNA complex was constructed in which two PrrAC molecules are bound to DNA in a symmetrical manner.
Figures


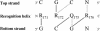

Similar articles
-
Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides.Biochemistry. 2006 Jun 27;45(25):7872-81. doi: 10.1021/bi060683g. Biochemistry. 2006. PMID: 16784239 Free PMC article.
-
Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box.J Mol Biol. 2000 Feb 4;295(5):1225-36. doi: 10.1006/jmbi.1999.3379. J Mol Biol. 2000. PMID: 10653699
-
Amino acid residues of RegA important for interactions with the CbbR-DNA complex of Rhodobacter sphaeroides.J Bacteriol. 2014 Sep;196(17):3179-90. doi: 10.1128/JB.01842-14. Epub 2014 Jun 23. J Bacteriol. 2014. PMID: 24957624 Free PMC article.
-
RegB/RegA, a global redox-responding two-component system.Adv Exp Med Biol. 2008;631:131-48. doi: 10.1007/978-0-387-78885-2_9. Adv Exp Med Biol. 2008. PMID: 18792686 Review.
-
Structure/function relationships in OmpR and other winged-helix transcription factors.Curr Opin Microbiol. 2002 Apr;5(2):135-41. doi: 10.1016/s1369-5274(02)00310-7. Curr Opin Microbiol. 2002. PMID: 11934608 Review.
Cited by
-
An integrated approach to reconstructing genome-scale transcriptional regulatory networks.PLoS Comput Biol. 2015 Feb 27;11(2):e1004103. doi: 10.1371/journal.pcbi.1004103. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25723545 Free PMC article.
-
Novel sequence-based method for identifying transcription factor binding sites in prokaryotic genomes.Bioinformatics. 2010 Nov 1;26(21):2672-7. doi: 10.1093/bioinformatics/btq501. Epub 2010 Aug 31. Bioinformatics. 2010. PMID: 20807838 Free PMC article.
-
Global analysis of photosynthesis transcriptional regulatory networks.PLoS Genet. 2014 Dec 11;10(12):e1004837. doi: 10.1371/journal.pgen.1004837. eCollection 2014 Dec. PLoS Genet. 2014. PMID: 25503406 Free PMC article.
-
Activation of the global gene regulator PrrA (RegA) from Rhodobacter sphaeroides.Biochemistry. 2006 Jun 27;45(25):7872-81. doi: 10.1021/bi060683g. Biochemistry. 2006. PMID: 16784239 Free PMC article.
-
The adherence-associated Fdp fasciclin I domain protein of the biohydrogen producer Rhodobacter sphaeroides is regulated by the global Prr pathway.Int J Hydrogen Energy. 2020 Oct 16;45(51):26840-26854. doi: 10.1016/j.ijhydene.2020.07.108. Int J Hydrogen Energy. 2020. PMID: 33093750 Free PMC article.
References
-
- Swem L.R., Elsen,S., Bird,T.H., Swem,D.L., Koch,H.G., Myllykallio,H., Daldal,F. and Bauer,C.E. (2001) The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol., 309, 121–138. - PubMed
-
- Robinson V.L., Buckler,D.R. and Stock,A.M. (2000) A tale of two components: a novel kinase and a regulatory switch. Nature Struct. Biol., 7, 626–633. - PubMed
-
- Masuda S., Matsumoto,Y., Nagashima,K.V., Shimada,K., Inoue,K., Bauer,C.E. and Matsuura,K. (1999) Structural and functional analyses of photosynthetic regulatory genes regA and regB from Rhodovulum sulfidophilum, Roseobacter denitrificans and Rhodobacter capsulatus. J. Bacteriol., 181, 4205–4215. - PMC - PubMed
-
- Emmerich R., Hennecke,H. and Fischer,H.M. (2000) Evidence for a functional similarity between the two-component regulatory systems RegSR, ActSR and RegBA (PrrBA) in alpha-Proteobacteria. Arch. Microbiol., 174, 307–313. - PubMed
-
- Birck C., Mourey,L., Gouet,P., Fabry,B., Schumacher,J., Rousseau,P., Kahn,D. and Samama,J.P. (1999) Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure Fold Des., 7, 1505–1515. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

