SVA elements are nonautonomous retrotransposons that cause disease in humans
- PMID: 14628287
- PMCID: PMC1180407
- DOI: 10.1086/380207
SVA elements are nonautonomous retrotransposons that cause disease in humans
Abstract
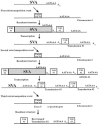
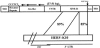
L1 elements are the only active autonomous retrotransposons in the human genome. The nonautonomous Alu elements, as well as processed pseudogenes, are retrotransposed by the L1 retrotransposition proteins working in trans. Here, we describe another repetitive sequence in the human genome, the SVA element. Our analysis reveals that SVA elements are currently active in the human genome. SVA elements, like Alus and L1s, occasionally insert into genes and cause disease. Furthermore, SVA elements are probably mobilized in trans by active L1 elements.
Figures

References
Electronic-Database Information
-
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for contigs AC037439 and AC068352 and BAC clone RP11-166N6 [contig AC016142])
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

