Template boundary definition in mammalian telomerase
- PMID: 14630939
- PMCID: PMC280623
- DOI: 10.1101/gad.1140303
Template boundary definition in mammalian telomerase
Abstract
Telomerase uses a short template sequence in its intrinsic RNA component to synthesize telomere repeats. Disruption of the helix P1b in human telomerase RNA or alteration of its distance from the template resulted in telomerase copying residues past the normal template boundary both in vivo and in vitro. Therefore, helix P1b is important for template boundary definition in human telomerase. Mouse telomerase RNA lacks helix P1b, and the boundary is established at 2 nt downstream of the 5'-end. The divergent structure of boundary definition elements in mammals, yeast, and ciliates suggests diverse mechanisms for template boundary definition in telomerase.
Figures
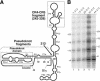



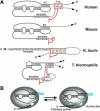
References
-
- Autexier C. and Greider, C.W. 1995. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & Dev. 9: 2227-2239. - PubMed
-
- Avilion A.A. 1995. Characterization and expression of human telomerase. In Graduate Program in Cellular and Developmental Biology, pp. 234. State University of New York at Stony Brook, Stony Brook, NY.
-
- Beattie T.L., Zhou, W., Robinson, M.O., and Harrington, L. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8: 177-180. - PubMed
-
- Blasco M.A., Lee, H.W., Hande, M.P., Samper, E., Lansdorp, P.M., DePinho, R.A., and Greider, C.W. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25-34. - PubMed
-
- Bryan T.M., Goodrich, K.J., and Cech, T.R. 2000. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6: 493-499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
