Fragile site orthologs FHIT/FRA3B and Fhit/Fra14A2: evolutionarily conserved but highly recombinogenic
- PMID: 14630947
- PMCID: PMC299872
- DOI: 10.1073/pnas.2336256100
Fragile site orthologs FHIT/FRA3B and Fhit/Fra14A2: evolutionarily conserved but highly recombinogenic
Abstract
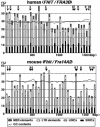
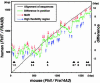
Common fragile sites are regions that show elevated susceptibility to DNA damage, leading to alterations that can contribute to cancer development. FRA3B, located at chromosome region 3p14.2, is the most frequently expressed human common fragile site, and allelic losses at FRA3B have been observed in many types of cancer. The FHIT gene, encompassing the FRA3B region, is a tumor-suppressor gene. To identify the features of FHIT/FRA3B that might contribute to fragility, sequences of the human FHIT and the flanking PTPRG gene were compared with those of murine Fhit and Ptprg. Human and mouse orthologous genes, FHIT and Fhit, are more highly conserved through evolution than PTPRG/Ptprg and yet contain more sequence elements that are exquisitely sensitive to genomic rearrangements, such as high-flexibility regions and long interspersed nuclear element 1s, suggesting that common fragile sites serve a function. The conserved AT-rich high-flexibility regions are the most characteristic of common fragile sites.
Figures


Similar articles
-
Sequence conservation at human and mouse orthologous common fragile regions, FRA3B/FHIT and Fra14A2/Fhit.Proc Natl Acad Sci U S A. 2001 May 8;98(10):5722-7. doi: 10.1073/pnas.091095898. Epub 2001 Apr 24. Proc Natl Acad Sci U S A. 2001. PMID: 11320209 Free PMC article.
-
Precise localization of the FHIT gene to the common fragile site at 3p14.2 (FRA3B) and characterization of homozygous deletions within FRA3B that affect FHIT transcription in tumor cell lines.Genes Chromosomes Cancer. 1997 Sep;20(1):16-23. doi: 10.1002/(sici)1098-2264(199709)20:1<16::aid-gcc3>3.0.co;2-c. Genes Chromosomes Cancer. 1997. PMID: 9290949
-
The murine Fhit gene is highly similar to its human orthologue and maps to a common fragile site region.Cancer Res. 1998 Aug 1;58(15):3409-14. Cancer Res. 1998. PMID: 9699673
-
The role of the FHIT/FRA3B locus in cancer.Annu Rev Genet. 1998;32:7-31. doi: 10.1146/annurev.genet.32.1.7. Annu Rev Genet. 1998. PMID: 9928473 Review.
-
An FHIT tumor suppressor gene?Genes Chromosomes Cancer. 1998 Apr;21(4):281-9. doi: 10.1002/(sici)1098-2264(199804)21:4<281::aid-gcc1>3.0.co;2-v. Genes Chromosomes Cancer. 1998. PMID: 9559339 Review.
Cited by
-
A Fhit-mimetic peptide suppresses annexin A4-mediated chemoresistance to paclitaxel in lung cancer cells.Oncotarget. 2016 May 24;7(21):29927-36. doi: 10.18632/oncotarget.9179. Oncotarget. 2016. PMID: 27166255 Free PMC article.
-
Quantifying the mechanisms for segmental duplications in mammalian genomes by statistical analysis and modeling.Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):4051-6. doi: 10.1073/pnas.0407957102. Epub 2005 Mar 1. Proc Natl Acad Sci U S A. 2005. PMID: 15741274 Free PMC article.
-
Fhit is a physiological target of the protein kinase Src.Proc Natl Acad Sci U S A. 2004 Mar 16;101(11):3775-9. doi: 10.1073/pnas.0400481101. Epub 2004 Mar 8. Proc Natl Acad Sci U S A. 2004. PMID: 15007172 Free PMC article.
-
BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function.Mol Cell Biol. 2004 Aug;24(15):6701-9. doi: 10.1128/MCB.24.15.6701-6709.2004. Mol Cell Biol. 2004. PMID: 15254237 Free PMC article.
-
Fhit delocalizes annexin a4 from plasma membrane to cytosol and sensitizes lung cancer cells to paclitaxel.PLoS One. 2013 Nov 6;8(11):e78610. doi: 10.1371/journal.pone.0078610. eCollection 2013. PLoS One. 2013. PMID: 24223161 Free PMC article.
References
-
- Glover, T. W., Berger, C., Coyle, J. & Echo, B. (1984) Hum. Genet. 67, 136-142. - PubMed
-
- Jones, C., Penny, L., Mattina, T., Yu, S., Baker, E., Voullaire, L., Langdon, W. Y., Sutherland, G. R., Richards, R. I. & Tunnacliffe, A. (1995) Nature 376, 145-149. - PubMed
-
- Hewett, D. R., Handt, O., Hobson, L., Mangelsdorf. M., Eyre, H. J., Baker, E., Sutherland, G. R., Schuffenhauer, S., Mao, J. I. & Richards, R. I. (1998) Mol. Cell 1, 773-781. - PubMed
-
- Yu, S., Mangelsdorf, M., Hewett, D., Hobson, L., Baker, E., Eyre, H. J., Lapsys, N., Le Paslier, D., Doggett, N. A., Sutherland, G. R., et al. (1997) Cell 88, 367-374. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

