Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif
- PMID: 14630973
- PMCID: PMC282827
- DOI: 10.1105/tpc.017541
Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif
Abstract
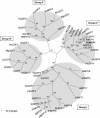
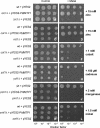
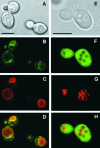
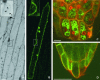
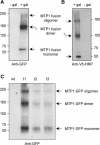
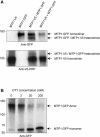
Cation diffusion facilitator (CDF) proteins are a recently discovered family of cation efflux transporters that might play an essential role in metal homeostasis and tolerance. Here, we describe the identification, characterization, and localization of PtdMTP1, a member of the CDF family from the hybrid poplar Populus trichocarpa x Populus deltoides. PtdMTP1 is expressed constitutively and ubiquitously, although at low levels. Heterologous expression in yeast showed that PtdMTP1 was able to complement the hypersensitivity of mutant strains to Zn but not to other metals, including Cd, Co, Mn, and Ni. PtdMTP1 fused to green fluorescent protein localized to the vacuolar membrane both in yeast and in plant cells, consistent with a function of PtdMTP1 in zinc sequestration. Overexpression of PtdMTP1 in Arabidopsis confers Zn tolerance. We show that PtdMTP1, when expressed in yeast and Arabidopsis, forms homooligomers, a novel feature of CDF members. Oligomer formation is disrupted by reducing agents, indicating possible disulfide bridge formation. PtdMTP1 also contains a conserved Leu zipper motif. Although not necessary for oligomer formation, Leu residues within this motif are required for PtdMTP1 functional activity.
Figures





References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. - PubMed
-
- Bachhawat, A.K., Manolson, M.F., Murdock, D.G., Garman, J.D., and Jones, E.W. (1993). The VPH2 gene encodes a 25 kDa protein required for activity of the yeast vacuolar H+-ATPase. Yeast 9, 175–184. - PubMed
-
- Bloss, T., Clemens, S., and Nies, D.H. (2002). Characterization of the ZAT1p zinc transporter from Arabidopsis thaliana in microbial model organisms and reconstituted proteoliposomes. Planta 214, 783–791. - PubMed
-
- Clemens, S. (2001). Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

