Ribosomal protection proteins and their mechanism of tetracycline resistance
- PMID: 14638464
- PMCID: PMC296194
- DOI: 10.1128/AAC.47.12.3675-3681.2003
Ribosomal protection proteins and their mechanism of tetracycline resistance
Figures
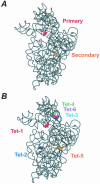

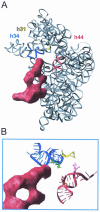
References
-
- Agrawal, R. K., A. B. Heagle, P. Penczek, R. A. Grassucci, and J. Frank. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6:643-647. - PubMed
-
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. - PubMed
-
- Brodersen, D. E., W. M. Clemons, A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. - PubMed
-
- Burdett, V. 1991. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J. Biol. Chem. 266:2872-2877. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

