Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition
- PMID: 14638858
- PMCID: PMC2173678
- DOI: 10.1083/jcb.200308020
Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition
Abstract
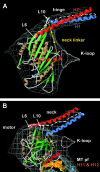
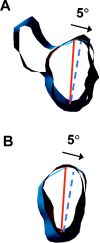
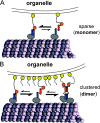
Caenhorhabditis elegans Unc104 kinesin transports synaptic vesicles at rapid velocities. Unc104 is primarily monomeric in solution, but recent motility studies suggest that it may dimerize when concentrated on membranes. Using cryo-electron microscopy, we observe two conformations of microtubule-bound Unc104: a monomeric state in which the two neck helices form an intramolecular, parallel coiled coil; and a dimeric state in which the neck helices form an intermolecular coiled coil. The intramolecular folded conformation is abolished by deletion of a flexible hinge separating the neck helices, indicating that it acts as a spacer to accommodate the parallel coiled-coil configuration. The neck hinge deletion mutation does not alter motor velocity in vitro but produces a severe uncoordinated phenotype in transgenic C. elegans, suggesting that the folded conformation plays an important role in motor regulation. We suggest that the Unc104 neck regulates motility by switching from a self-folded, repressed state to a dimerized conformation that can support fast processive movement.
Figures




Comment in
-
Breathing down the neck of Unc104.J Cell Biol. 2003 Nov 24;163(4):693-5. doi: 10.1083/jcb.200310137. J Cell Biol. 2003. PMID: 14638852 Free PMC article. Review.
References
-
- Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221–241. - PubMed
-
- Bloom, G.S. 2001. The UNC-104/KIF1 family of kinesins. Curr. Opin. Cell Biol. 13:36–40. - PubMed
-
- Burkhard, P., R.A. Kammerer, M.O. Steinmetz, G.P. Bourenkov, and U. Aebi. 2000. The coiled coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure Fold. Des. 8:223–230. - PubMed

