Mammalian GGAs act together to sort mannose 6-phosphate receptors
- PMID: 14638859
- PMCID: PMC2173681
- DOI: 10.1083/jcb.200308038
Mammalian GGAs act together to sort mannose 6-phosphate receptors
Abstract
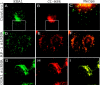
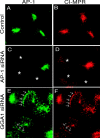
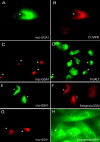
The GGAs (Golgi-localized, gamma ear-containing, ADP ribosylation factor-binding proteins) are multidomain proteins implicated in protein trafficking between the Golgi and endosomes. We examined whether the three mammalian GGAs act independently or together to mediate their functions. Using cryo-immunogold electron microscopy, the three GGAs were shown to colocalize within coated buds and vesicles at the trans-Golgi network (TGN) of HeLa cells. In vitro binding experiments revealed multidomain interactions between the GGAs, and chemical cross-linking experiments demonstrated that GGAs 1 and 2 form a complex on Golgi membranes. RNA interference of each GGA resulted in decreased levels of the other GGAs and their redistribution from the TGN to cytosol. This was associated with impaired incorporation of the cation-independent mannose 6-phosphate receptor into clathrin-coated vesicles at the TGN, partial redistribution of the receptor to endosomes, and missorting of cathepsin D. The morphology of the TGN was also altered. These findings indicate that the three mammalian GGAs cooperate to sort cargo and are required for maintenance of TGN structure.
Figures







Similar articles
-
ArfGAP3 regulates the transport of cation-independent mannose 6-phosphate receptor in the post-Golgi compartment.Curr Biol. 2013 Oct 7;23(19):1945-51. doi: 10.1016/j.cub.2013.07.087. Epub 2013 Sep 26. Curr Biol. 2013. PMID: 24076238 Free PMC article.
-
Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network.Science. 2002 Sep 6;297(5587):1700-3. doi: 10.1126/science.1075327. Science. 2002. PMID: 12215646
-
The GGA proteins: key players in protein sorting at the trans-Golgi network.Eur J Cell Biol. 2004 Jul;83(6):257-62. doi: 10.1078/0171-9335-00374. Eur J Cell Biol. 2004. PMID: 15511083 Review.
-
The trans-Golgi network accessory protein p56 promotes long-range movement of GGA/clathrin-containing transport carriers and lysosomal enzyme sorting.Mol Biol Cell. 2007 Sep;18(9):3486-501. doi: 10.1091/mbc.e07-02-0190. Epub 2007 Jun 27. Mol Biol Cell. 2007. PMID: 17596511 Free PMC article.
-
The structure and function of GGAs, the traffic controllers at the TGN sorting crossroads.Cell Struct Funct. 2003 Oct;28(5):431-42. doi: 10.1247/csf.28.431. Cell Struct Funct. 2003. PMID: 14745135 Review.
Cited by
-
HIV-1 buds predominantly at the plasma membrane of primary human macrophages.PLoS Pathog. 2007 Mar;3(3):e36. doi: 10.1371/journal.ppat.0030036. PLoS Pathog. 2007. PMID: 17381240 Free PMC article.
-
Protein sorting from endosomes to the TGN.Front Cell Dev Biol. 2023 Feb 21;11:1140605. doi: 10.3389/fcell.2023.1140605. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36895788 Free PMC article. Review.
-
The Biology of Lysosomes: From Order to Disorder.Biomedicines. 2023 Jan 14;11(1):213. doi: 10.3390/biomedicines11010213. Biomedicines. 2023. PMID: 36672721 Free PMC article. Review.
-
Drosophila GGA model: an ultimate gateway to GGA analysis.Traffic. 2011 Dec;12(12):1821-38. doi: 10.1111/j.1600-0854.2011.01285.x. Epub 2011 Oct 13. Traffic. 2011. PMID: 21923734 Free PMC article.
-
GGA1 interacts with the endosomal Na+/H+ exchanger NHE6 governing localization to the endosome compartment.J Biol Chem. 2024 Aug;300(8):107552. doi: 10.1016/j.jbc.2024.107552. Epub 2024 Jul 11. J Biol Chem. 2024. PMID: 39002678 Free PMC article.
References
-
- Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

