Structure of sphingomyelin bilayers: a simulation study
- PMID: 14645055
- PMCID: PMC1303667
- DOI: 10.1016/S0006-3495(03)74780-8
Structure of sphingomyelin bilayers: a simulation study
Abstract
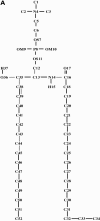
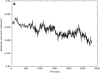
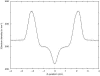
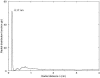
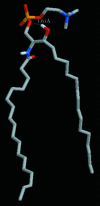
We have carried out a molecular dynamics simulation of a hydrated 18:0 sphingomyelin lipid bilayer. The bilayer contained 1600 sphingomyelin (SM) molecules, and 50,592 water molecules. After construction and initial equilibration, the simulation was run for 3.8 ns at a constant temperature of 50 degrees C and a constant pressure of 1 atm. We present properties of the bilayer calculated from the simulation, and compare with experimental data and with properties of dipalmitoyl phosphatidylcholine (DPPC) bilayers. The SM bilayers are significantly more ordered and compact than DPPC bilayers at the same temperature. SM bilayers also exhibit significant intramolecular hydrogen bonding between phosphate ester oxygen and hydroxyl hydrogen atoms. This results in a decreased hydration in the polar region of the SM bilayer compared with DPPC. Since our simulation system is very large we have calculated the power spectrum of bilayer undulation and peristaltic modes, and we compare these data with similar calculations for DPPC bilayers. We find that the SM bilayer has significantly larger bending modulus and area compressibility compared to DPPC.
Figures







References
-
- Ahmed, S. N., D. A. Brown, and E. London. 1997. On the origin of sphingolipid/cholesterol rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble liquid-ordered lipid phase in model membranes. Biochemistry. 36:10944–10953. - PubMed
-
- Allen, M. P., and D. J. Tildesley. 1987. Computer Simulation of Liquids. Clarendon Press, Oxford, UK.
-
- Aman, M. J., and K. S. Ravichandran. 2000. A requirement for lipid rafts in B-cell receptor induced Ca2+ flux. Curr. Biol. 10:393–396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

