Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa
- PMID: 14645283
- PMCID: PMC296264
- DOI: 10.1128/JB.185.24.7222-7230.2003
Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa
Abstract
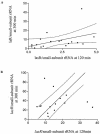
Quorum sensing (QS)-based transcriptional responses in Pseudomonas aeruginosa have been defined on the basis of increases in transcript levels of QS-controlled genes such as lasB and aprA following the hierarchical transcriptional increases of central controllers such as the lasR gene. These increases occur at high bacterial concentrations such as early-stationary-phase growth in vitro. However, the extent to which the increases occur in a variety of clinical and environmental isolates has not been determined nor is there extensive information on allelic variation in lasR genes. An analysis of the sequences of the lasR gene among 66 clinical and environmental isolates showed that 81% have a sequence either identical to that of strain PAO1 or with a silent mutation, 15% have nucleotide changes resulting in amino acid changes, and 5% have an insertion sequence in the lasR gene. Using real-time PCR to quantify transcript levels of lasR, lasB, and aprA in the early log and early stationary phases among 35 isolates from bacteremia and pneumonia cases and the environment, we found most (33 of 35) strains had increases in lasR transcripts in early stationary phase but with a very wide range of final transcript levels per cell. There was a strong correlation (r(2) = 0.84) between early-log- and early-stationary-phase transcript levels in all strains, but this finding remained true only for the 50% of strains above the median level of lasR found in early log phase. There were significant (P < 0.05) but weak-to-modest correlations of lasR transcript levels with aprA (r(2) = 0.2) and lasB (r(2) = 0.5) transcript levels, but again this correlation occurred only in the 50% of P. aeruginosa strains with the highest levels of lasR transcripts in early stationary phase. There were no differences in distribution of lasR alleles among the bacteremia, pneumonia, or environmental isolates. Overall, only about 50% of P. aeruginosa strains from clinical and environmental sources show a lasR-dependent increase in the transcription of aprA and lasB genes, indicating that for about 50% of clinical isolates this regulatory system may not play a significant role in pathogenesis.
Figures

References
-
- Bonten, M. J., D. C. Bergmans, A. W. Ambergen, P. W. de Leeuw, S. van der Geest, E. E. Stobberingh, and C. A. Gaillard. 1996. Risk factors for pneumonia, and colonization of respiratory tract and stomach in mechanically ventilated ICU patients. Am. J. Respir. Crit. Care Med. 154:1339-1346. - PubMed
-
- Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

