Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature
- PMID: 14645288
- PMCID: PMC296247
- DOI: 10.1128/JB.185.24.7266-7272.2003
Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature
Abstract
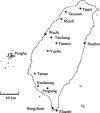
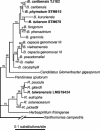
Following the initial discovery of two legume-nodulating Burkholderia strains (L. Moulin, A. Munive, B. Dreyfus, and C. Boivin-Masson, Nature 411:948-950, 2001), we identified as nitrogen-fixing legume symbionts at least 50 different strains of Burkholderia caribensis and Ralstonia taiwanensis, all belonging to the beta-subclass of proteobacteria, thus extending the phylogenetic diversity of the rhizobia. R. taiwanensis was found to represent 93% of the Mimosa isolates in Taiwan, indicating that beta-proteobacteria can be the specific symbionts of a legume. The nod genes of rhizobial beta-proteobacteria (beta-rhizobia) are very similar to those of rhizobia from the alpha-subclass (alpha-rhizobia), strongly supporting the hypothesis of the unique origin of common nod genes. The beta-rhizobial nod genes are located on a 0.5-Mb plasmid, together with the nifH gene, in R. taiwanensis and Burkholderia phymatum. Phylogenetic analysis of available nodA gene sequences clustered beta-rhizobial sequences in two nodA lineages intertwined with alpha-rhizobial sequences. On the other hand, the beta-rhizobia were grouped with free-living nitrogen-fixing beta-proteobacteria on the basis of the nifH phylogenetic tree. These findings suggest that beta-rhizobia evolved from diazotrophs through multiple lateral nod gene transfers.
Figures




References
-
- Chen, W. M., S. Laevens, T. M. Lee, T. Coenye, P. de Vos, M. Mergeay, and P. Vandamme. 2001. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51:1729-1735. - PubMed
-
- W.-M. Chen, E. K. James, A. R. Prescott, M. Kierans, and J. I. Sprent. Nodulation of Mimosa spp. by the β-proteobacterium Ralstonia taiwanensis. Mol. Plant-Microbe Interact., in press. - PubMed
-
- Deschodt, C. C., and B. W. Strijdom. 1976. Effective nodulation of Aspalathus linearis by rhizobia from other Aspalathus species. Phytophylactica 8:103-104.
-
- Fani, R., R. Gallo, and P. Lio. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51:1-11. - PubMed
-
- Felsenstein, J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27:401-410.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

