Ubiquitin depletion as a key mediator of toxicity by translational inhibitors
- PMID: 14645527
- PMCID: PMC309641
- DOI: 10.1128/MCB.23.24.9251-9261.2003
Ubiquitin depletion as a key mediator of toxicity by translational inhibitors
Abstract
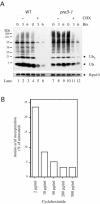
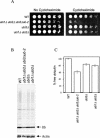
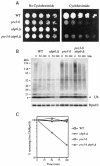
Cycloheximide acts at the large subunit of the ribosome to inhibit translation. Here we report that ubiquitin levels are critical for the survival of Saccharomyces cerevisiae cells in the presence of cycloheximide: ubiquitin overexpression confers resistance to cycloheximide, while a reduced ubiquitin level confers sensitivity. Consistent with these findings, ubiquitin is unstable in yeast (t(1/2) = 2 h) and is rapidly depleted upon cycloheximide treatment. Cycloheximide does not noticeably enhance ubiquitin turnover, but serves principally to block ubiquitin synthesis. Cycloheximide also induces UBI4, the polyubiquitin gene. The cycloheximide-resistant phenotype of ubiquitin overexpressors is also characteristic of partial-loss-of-function proteasome mutants. Ubiquitin is stabilized in these mutants, which may account for their cycloheximide resistance. Previous studies have reported that ubiquitin is destabilized in the absence of Ubp6, a proteasome-associated deubiquitinating enzyme, and that ubp6 mutants are hypersensitive to cycloheximide. Consistent with the model that cycloheximide-treated cells are ubiquitin deficient, the cycloheximide sensitivity of ubp6 mutants can be rescued either by ubiquitin overexpression or by mutations in proteasome subunit genes. These results also show that ubiquitin wasting in ubp6 mutants is proteasome mediated. Ubiquitin overexpression rescued cells from additional translational inhibitors such as anisomycin and hygromycin B, suggesting that ubiquitin depletion may constitute a widespread mechanism for the toxicity of translational inhibitors.
Figures


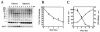

References
-
- Abraham, A. K., and A. Pihl. 1983. Effect of protein synthesis inhibitors on the fidelity of translation in eukaryotic systems. Biochim. Biophys. Acta 741:197-203. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1994. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
-
- Brostrom, C. O., and M. A. Brostrom. 1998. Regulation of translational initiation during cellular responses to stress. Prog. Nucleic Acid Res. Mol. Biol. 58:79-125. - PubMed
-
- Burbea, M., L. Dreier, J. S. Dittman, M. E. Grunwald, and J. Kaplan. 2002. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron 35:107-120. - PubMed
-
- Chen, Y., and P. W. Piper. 1995. Consequences of overexpression of ubiquitin in yeast: elevated tolerances of osmostress, ethanol and canavanine, yet reduced tolerances of cadmium, arsenite and paromomycin. Biochim. Biophys. Acta 1268:59-64. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
