The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork
- PMID: 14645529
- PMCID: PMC309713
- DOI: 10.1128/MCB.23.24.9178-9188.2003
The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork
Abstract
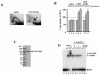
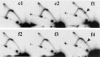
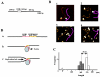
The replication fork barrier site (RFB) is an approximately 100-bp DNA sequence located near the 3' end of the rRNA genes in the yeast Saccharomyces cerevisiae. The gene FOB1 is required for this RFB activity. FOB1 is also necessary for recombination in the ribosomal DNA (rDNA), including increase and decrease of rDNA repeat copy number, production of extrachromosomal rDNA circles, and possibly homogenization of the repeats. Despite the central role that Foblp plays in both replication fork blocking and rDNA recombination, the molecular mechanism by which Fob1p mediates these activities has not been determined. Here, I show by using chromatin immunoprecipitation, gel shift, footprinting, and atomic force microscopy assays that Fob1p directly binds to the RFB. Fob1p binds to two separated sequences in the RFB. A predicted zinc finger motif in Fob1p was shown to be essential for the RFB binding, replication fork blocking, and rDNA recombination activities. The RFB seems to wrap around Fob1p, and this wrapping structure may be important for function in the rDNA repeats.
Figures




References
-
- Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. - PubMed
-
- Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. - PubMed
-
- Brewer, B. J., D. Lockshon, and W. L. Fangman. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71:267-276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
