Affinity purification of specific chromatin segments from chromosomal loci in yeast
- PMID: 14645537
- PMCID: PMC309680
- DOI: 10.1128/MCB.23.24.9275-9282.2003
Affinity purification of specific chromatin segments from chromosomal loci in yeast
Abstract
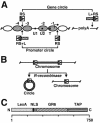
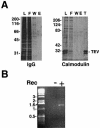
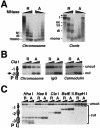
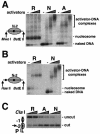
Single-copy gene and promoter regions have been excised from yeast chromosomes and have been purified as chromatin by conventional and affinity methods. Promoter regions isolated in transcriptionally repressed and activated states maintain their characteristic chromatin structures. Gel filtration analysis establishes the uniformity of the transcriptionally activated state. Activator proteins interact in the manner anticipated from previous studies in vivo. This work opens the way to the direct study of specific gene regions of eukaryotic chromosomes in diverse functional and structural states.
Figures


References
-
- Ansari, A., T. H. Cheng, and M. R. Gartenberg. 1999. Isolation of selected chromatin fragments from yeast by site-specific recombination in vivo. Methods 17:104-111. - PubMed
-
- Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 11:1587-1598. - PubMed
-
- Godde, J. S., and A. P. Wolffe. 1995. Disruption of reconstituted nucleosomes. The effect of particle concentration, MgCl2 and KCl concentration, the histone tails, and temperature. J. Biol. Chem. 270:27399-27402. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
