Novel system for the simultaneous analysis of geminivirus DNA replication and plant interactions in Nicotiana benthamiana
- PMID: 14645587
- PMCID: PMC296063
- DOI: 10.1128/jvi.77.24.13315-13322.2003
Novel system for the simultaneous analysis of geminivirus DNA replication and plant interactions in Nicotiana benthamiana
Abstract
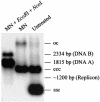
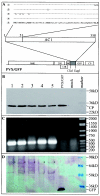
The origin of replication of African cassava mosaic virus (ACMV) and a gene expression vector based on Potato virus X were exploited to devise an in planta system for functional analysis of the geminivirus replication-associated protein (Rep) in transgenic Nicotiana benthamiana line pOri-2. This line contains an integrated copy of a tandem repeat of the ACMV origin of replication flanking nonviral sequences that can be mobilized and replicated by Rep as an episomal replicon. A Rep-GFP fusion protein can also mobilize and amplify the replicon, facilitating Rep detection in planta. The activity of Rep and its mutants, Rep-mediated host response, and the correlation between Rep intracellular localization and biological functions could be effectively assessed by using this in planta system. Our results indicate that modification of amino acid residues R(2), R(5), R(7) and K(11) or H(56), L(57) and H(58) prevent Rep function in replication. This defect correlates with possible loss of Rep nuclear localization and inability to trigger the host defense mechanism resembling a hypersensitive response.
Figures





References
-
- Argüello-Astorga, G. R., R. G. Guevara-González, L. R. Herrera-Estrella, and R. F. Rivera-Bustamante. 1994. Geminivirus replication origins have a group-specific organization of iterative elements: a model for replication. Virology 203:90-100. - PubMed
-
- Berzal-Herranz, A., A. De La Cruz, F. Tenllado, J. R. Diaz-Ruiz, L. Lopez, A. I. Sanz, C. Vaquero, M. T. Serra, and I. Garcia-Luque. 1994. The Capsicum L3 gene-mediated resistance against tobamoviruses is elicited by the coat protein. Virology 209:498-505. - PubMed
-
- Briddon, R. W., J. Watts, P. G. Markham, and J. Stanley. 1989. The coat protein of beet curly top virus is essential for infectivity. Virology 172:628-633. - PubMed
-
- Chapman, S., T. Kavanagh, and D. Baulcombe. 1992. Potato virus X as a vector for gene expression in plants. Plant J. 2:549-557. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

