De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions
- PMID: 14645602
- PMCID: PMC296083
- DOI: 10.1128/jvi.77.24.13439-13447.2003
De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions
Abstract
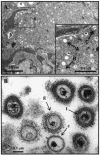
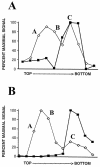
Rhesus monkey rhadinovirus (RRV) is one of the closest phylogenetic relatives to the human pathogen Kaposi's sarcoma-associated herpesvirus (KSHV), yet it has the distinct experimental advantage of entering efficiently into lytic replication and growing to high titers in culture. RRV therefore holds promise as a potentially attractive model with which to study gammaherpesvirus structure and assembly. We have isolated RRV capsids, determined their molecular composition, and identified the genes encoding five of the main capsid structural proteins. Our data indicate that, as with other herpesviruses, lytic infection with RRV leads to the synthesis of three distinct intranuclear capsid species. However, in contrast to the inefficiency of KSHV maturation following reactivation from latently infected B-cell lines (K. Nealon, W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes, J. Virol. 75:2866-2878, 2001), de novo infection of immortalized rhesus fibroblasts with RRV results in the release of high levels of infectious virions with genome-containing C capsids at their center. Together, our findings argue for the use of RRV as a powerful model with which to study the structure and assembly of gammaherpesviruses and, specifically, the human rhadinovirus,KSHV.
Figures




Similar articles
-
Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging.J Virol. 2003 Dec;77(24):13182-93. doi: 10.1128/jvi.77.24.13182-13193.2003. J Virol. 2003. PMID: 14645575 Free PMC article.
-
Maturation and vesicle-mediated egress of primate gammaherpesvirus rhesus monkey rhadinovirus require inner tegument protein ORF52.J Virol. 2014 Aug;88(16):9111-28. doi: 10.1128/JVI.01502-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899183 Free PMC article.
-
Rhesus monkey rhadinovirus: a model for the study of KSHV.Curr Top Microbiol Immunol. 2007;312:43-69. doi: 10.1007/978-3-540-34344-8_2. Curr Top Microbiol Immunol. 2007. PMID: 17089793 Review.
-
A Conserved Leucine Zipper Motif in Gammaherpesvirus ORF52 Is Critical for Distinct Microtubule Rearrangements.J Virol. 2017 Aug 10;91(17):e00304-17. doi: 10.1128/JVI.00304-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615210 Free PMC article.
-
Rhesus macaque rhadinovirus-associated disease.Curr Opin Virol. 2013 Jun;3(3):245-50. doi: 10.1016/j.coviro.2013.05.016. Epub 2013 Jun 6. Curr Opin Virol. 2013. PMID: 23747119 Free PMC article. Review.
Cited by
-
KSHV targets multiple leukocyte lineages during long-term productive infection in NOD/SCID mice.J Clin Invest. 2006 Jul;116(7):1963-73. doi: 10.1172/JCI27249. Epub 2006 Jun 22. J Clin Invest. 2006. PMID: 16794734 Free PMC article.
-
Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging.J Virol. 2003 Dec;77(24):13182-93. doi: 10.1128/jvi.77.24.13182-13193.2003. J Virol. 2003. PMID: 14645575 Free PMC article.
-
Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization.Structure. 2014 Oct 7;22(10):1385-98. doi: 10.1016/j.str.2014.05.019. Epub 2014 Sep 11. Structure. 2014. PMID: 25220471 Free PMC article.
-
Mutational analysis of the herpes simplex virus triplex protein VP19C.J Virol. 2006 Feb;80(3):1537-48. doi: 10.1128/JVI.80.3.1537-1548.2006. J Virol. 2006. PMID: 16415029 Free PMC article.
-
Proteomic characterization of bovine herpesvirus 4 extracellular virions.J Virol. 2012 Nov;86(21):11567-80. doi: 10.1128/JVI.00456-12. Epub 2012 Aug 15. J Virol. 2012. PMID: 22896609 Free PMC article.
References
-
- Ablashi, D. V., G. R. Armstrong, U. Heine, and R. A. Manaker. 1971. Propagation of Herpesvirus saimiri in human cells. J. Natl. Cancer Inst. 47:241-244. - PubMed
-
- Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. - PMC - PubMed
-
- Ambinder, R. F. 2001. Epstein-Barr virus associated lymphoproliferations in the AIDS setting. Eur. J. Cancer 37:1209-1216. - PubMed
-
- Beral, V. 1991. Epidemiology of Kaposi's sarcoma. Cancer Surv. 10:5-22. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

