Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome
- PMID: 14645721
- PMCID: PMC299976
- DOI: 10.1073/pnas.2435133100
Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome
Abstract
More than 50 leaf rust resistance (Lr) genes against the fungal pathogen Puccinia triticina have been identified in the wheat gene pool, and a large number of them have been extensively used in breeding. Of the 50 Lr genes, all are known only from their phenotype and/or map position except for Lr21, which was cloned recently. For many years, the problems of molecular work in the large (1.6 x 10(10) bp), highly repetitive (80%), and hexaploid bread wheat (Triticum aestivum L.) genome have hampered map-based cloning. Here, we report the isolation of the Lr gene Lr10 from hexaploid wheat by using a combination of subgenome map-based cloning and haplotype studies in the genus Triticum. Lr10 is a single-copy gene on chromosome 1AS. It encodes a CC-NBS-LRR type of protein with an N-terminal domain, which is under diversifying selection. When overexpressed in transgenic wheat plants, Lr10 confers enhanced resistance to leaf rust. Lr10 has similarities to RPM1 in Arabidopsis thaliana and to resistance gene analogs in rice and barley, but is not closely related to other wheat Lr genes based on Southern analysis. We conclude that map-based cloning of genes of agronomic importance in hexaploid wheat is now feasible, opening perspectives for molecular bread wheat improvement trough transgenic strategies and diagnostic allele detection.
Figures
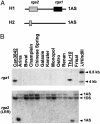
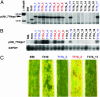

References
-
- Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826-833. - PubMed
-
- Hulbert, S. H., Webb, C. A., Smith, S. M. & Sun, Q. (2001) Annu. Rev. Phytopathol. 39, 285-312. - PubMed
-
- Buschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., vanDaelen, R., vanderLee, T., Diergaarde, P., Groenendijk, J., et al. (1997) Cell 88, 695-705. - PubMed
-
- Yu, Y., Tomkins, J. P., Waugh, R., Frisch, D. A., Kudrna, D., Kleinhofs, A., Brueggeman, R. S., Muehlbauer, G. J., Wise, R. P. & Wing, R. A. (2000) Theor. Appl. Genet. 101, 1093-1099.
-
- Halterman, D., Zhou, F. S., Wei, F. S., Wise, R. P. & Schulze-Lefert, P. (2001) Plant J. 25, 335-348. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

