Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses
- PMID: 14645724
- PMCID: PMC300730
- DOI: 10.1104/pp.103.025742
Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses
Abstract
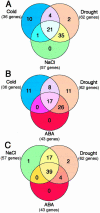
To identify cold-, drought-, high-salinity-, and/or abscisic acid (ABA)-inducible genes in rice (Oryza sativa), we prepared a rice cDNA microarray including about 1700 independent cDNAs derived from cDNA libraries prepared from drought-, cold-, and high-salinity-treated rice plants. We confirmed stress-inducible expression of the candidate genes selected by microarray analysis using RNA gel-blot analysis and finally identified a total of 73 genes as stress inducible including 58 novel unreported genes in rice. Among them, 36, 62, 57, and 43 genes were induced by cold, drought, high salinity, and ABA, respectively. We observed a strong association in the expression of stress-responsive genes and found 15 genes that responded to all four treatments. Venn diagram analysis revealed greater cross talk between signaling pathways for drought, ABA, and high-salinity stresses than between signaling pathways for cold and ABA stresses or cold and high-salinity stresses in rice. The rice genome database search enabled us not only to identify possible known cis-acting elements in the promoter regions of several stress-inducible genes but also to expect the existence of novel cis-acting elements involved in stress-responsive gene expression in rice stress-inducible promoters. Comparative analysis of Arabidopsis and rice showed that among the 73 stress-inducible rice genes, 51 already have been reported in Arabidopsis with similar function or gene name. Transcriptome analysis revealed novel stress-inducible genes, suggesting some differences between Arabidopsis and rice in their response to stress.
Figures



Comment in
-
Abiotic stress in rice. An "omic" approach.Plant Physiol. 2006 Apr;140(4):1139-41. doi: 10.1104/pp.104.900188. Plant Physiol. 2006. PMID: 16607027 Free PMC article. No abstract available.
Similar articles
-
Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray.Plant J. 2002 Aug;31(3):279-92. doi: 10.1046/j.1365-313x.2002.01359.x. Plant J. 2002. PMID: 12164808
-
Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray.Plant Cell. 2001 Jan;13(1):61-72. doi: 10.1105/tpc.13.1.61. Plant Cell. 2001. PMID: 11158529 Free PMC article.
-
A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice.Plant Biol (Stuttg). 2017 Mar;19(2):257-267. doi: 10.1111/plb.12514. Epub 2016 Nov 1. Plant Biol (Stuttg). 2017. PMID: 27718311
-
Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress.Plant Cell Rep. 2013 Jul;32(7):985-1006. doi: 10.1007/s00299-013-1414-5. Epub 2013 Mar 19. Plant Cell Rep. 2013. PMID: 23508256 Review.
-
ABA-dependent and ABA-independent signaling in response to osmotic stress in plants.Curr Opin Plant Biol. 2014 Oct;21:133-139. doi: 10.1016/j.pbi.2014.07.009. Epub 2014 Aug 9. Curr Opin Plant Biol. 2014. PMID: 25104049 Review.
Cited by
-
Upstream regulatory architecture of rice genes: summarizing the baseline towards genus-wide comparative analysis of regulatory networks and allele mining.Rice (N Y). 2015 Feb 28;8:14. doi: 10.1186/s12284-015-0041-x. eCollection 2015. Rice (N Y). 2015. PMID: 25844119 Free PMC article.
-
The Role of the Atypical Kinases ABC1K7 and ABC1K8 in Abscisic Acid Responses.Front Plant Sci. 2016 Mar 24;7:366. doi: 10.3389/fpls.2016.00366. eCollection 2016. Front Plant Sci. 2016. PMID: 27047531 Free PMC article.
-
Exogenous abscisic acid significantly affects proteome in tea plant (Camellia sinensis) exposed to drought stress.Hortic Res. 2014 Jun 25;1:14029. doi: 10.1038/hortres.2014.29. eCollection 2014. Hortic Res. 2014. PMID: 27076915 Free PMC article.
-
De novo sequencing and characterization of the floral transcriptome of Dendrocalamus latiflorus (Poaceae: Bambusoideae).PLoS One. 2012;7(8):e42082. doi: 10.1371/journal.pone.0042082. Epub 2012 Aug 14. PLoS One. 2012. PMID: 22916120 Free PMC article.
-
A shared response of thaumatin like protein, chitinase, and late embryogenesis abundant protein3 to environmental stresses in tea [Camellia sinensis (L.) O. Kuntze].Funct Integr Genomics. 2012 Aug;12(3):565-71. doi: 10.1007/s10142-012-0279-y. Epub 2012 Apr 29. Funct Integr Genomics. 2012. PMID: 22543414
References
-
- Aguan K, Sugawara K, Suzuki N, Kusano T (1991) Isolation of genes for low-temperature-induced proteins in rice by a simple subtractive method. Plant Cell Physiol 32: 1285-1289
-
- Bajaj S, Targolli J, Liu LF, Ho THD, Wu R (1999) Transgenic approaches to increase dehydration-stress tolerance in plants. Mol Breed 5: 493-503
-
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that control cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701-713 - PubMed
-
- Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2: 48-54
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

