The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules
- PMID: 14645732
- PMCID: PMC300743
- DOI: 10.1104/pp.103.031237
The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules
Abstract
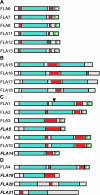
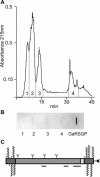
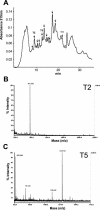
Fasciclin-like arabinogalactan proteins (FLAs) are a subclass of arabinogalactan proteins (AGPs) that have, in addition to predicted AGP-like glycosylated regions, putative cell adhesion domains known as fasciclin domains. In other eukaryotes (e.g. fruitfly [Drosophila melanogaster] and humans [Homo sapiens]), fasciclin domain-containing proteins are involved in cell adhesion. There are at least 21 FLAs in the annotated Arabidopsis genome. Despite the deduced proteins having low overall similarity, sequence analysis of the fasciclin domains in Arabidopsis FLAs identified two highly conserved regions that define this motif, suggesting that the cell adhesion function is conserved. We show that FLAs precipitate with beta-glucosyl Yariv reagent, indicating that they share structural characteristics with AGPs. Fourteen of the FLA family members are predicted to be C-terminally substituted with a glycosylphosphatidylinositol anchor, a cleavable form of membrane anchor for proteins, indicating different FLAs may have different developmental roles. Publicly available microarray and expressed sequence tag data were used to select FLAs for further expression analysis. RNA gel blots for a number of FLAs indicate that they are likely to be important during plant development and in response to abiotic stress. FLAs 1,2, and 8 show a rapid decrease in mRNA abundance in response to the phytohormone abscisic acid. Also, the accumulation of FLA1 and FLA2 transcripts differs during callus and shoot development, indicating that the proteins may be significant in the process of competence acquisition and induction of shoot development.
Figures

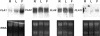

References
-
- AGI (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815 - PubMed
-
- Bacic A, Currie G, Gilson P, Mau S-L, Oxley D, Schultz CJ, Sommer-Knudsen J, Clarke AE (2000) Structural classes of arabinogalactan-proteins. In EA Nothnagel, A Bacic, AE Clarke, eds, Cell and Developmental Biology of Arabinogalactan-Proteins. Kluwer Academic/Plenum Publishers, New York, pp 11-23
-
- Bacic A, Harris PJ, Stone BA (1988) Structure and function of plant cell walls. In J Priess, ed, The Biochemistry of Plants, Vol 14. Academic Press, New York, pp 297-371
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

