Tracking EcoKI and DNA fifty years on: a golden story full of surprises
- PMID: 14654681
- PMCID: PMC291878
- DOI: 10.1093/nar/gkg944
Tracking EcoKI and DNA fifty years on: a golden story full of surprises
Abstract
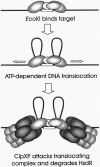
1953 was a historical year for biology, as it marked the birth of the DNA helix, but also a report by Bertani and Weigle on 'a barrier to infection' of bacteriophage lambda in its natural host, Escherichia coli K-12, that could be lifted by 'host-controlled variation' of the virus. This paper lay dormant till Nobel laureate Arber and PhD student Dussoix showed that the lambda DNA was rejected and degraded upon infection of different bacterial hosts, unless it carried host-specific modification of that DNA, thus laying the foundations for the phenomenon of restriction and modification (R-M). The restriction enzyme of E.coli K-12, EcoKI, was purified in 1968 and required S-adenosylmethionine (AdoMet) and ATP as cofactors. By the end of the decade there was substantial evidence for a chromosomal locus hsdK with three genes encoding restriction (R), modification (M) and specificity (S) subunits that assembled into a large complex of >400 kDa. The 1970s brought the message that EcoKI cut away from its DNA recognition target, to which site the enzyme remained bound while translocating the DNA past itself, with concomitant ATP hydrolysis and subsequent double-strand nicks. This translocation event created clearly visible DNA loops in the electron microscope. EcoKI became the archetypal Type I R-M enzyme with curious DNA translocating properties reminiscent of helicases, recognizing the bipartite asymmetric site AAC(N6)GTGC. Cloning of the hsdK locus in 1976 facilitated molecular understanding of this sophisticated R-M complex and in an elegant 'pas de deux' Murray and Dryden constructed the present model based on a large body of experimental data plus bioinformatics. This review celebrates the golden anniversary of EcoKI and ends with the exciting progress on the vital issue of restriction alleviation after DNA damage, also first reported in 1953, which involves intricate control of R subunit activity by the bacterial proteasome ClpXP, important results that will keep scientists on the EcoKI track for another 50 years to come.
Figures



Similar articles
-
Target recognition by EcoKI: the recognition domain is robust and restriction-deficiency commonly results from the proteolytic control of enzyme activity.J Mol Biol. 2001 Mar 30;307(3):951-63. doi: 10.1006/jmbi.2001.4543. J Mol Biol. 2001. PMID: 11273713
-
Sequence-specific DNA binding by EcoKI, a type IA DNA restriction enzyme.J Mol Biol. 1998 Nov 13;283(5):963-76. doi: 10.1006/jmbi.1998.2143. J Mol Biol. 1998. PMID: 9799636
-
Modulation of EcoKI restriction in vivo: role of the lambda Gam protein and plasmid metabolism.J Bacteriol. 1997 Mar;179(6):1852-6. doi: 10.1128/jb.179.6.1852-1856.1997. J Bacteriol. 1997. PMID: 9068628 Free PMC article.
-
Portraits of viruses: bacteriophage lambda.Intervirology. 1980;13(3):133-53. doi: 10.1159/000149119. Intervirology. 1980. PMID: 6246031 Review. No abstract available.
-
Bacteriophage lambda: the untold story.J Mol Biol. 1999 Oct 22;293(2):177-80. doi: 10.1006/jmbi.1999.3137. J Mol Biol. 1999. PMID: 10550203 Review.
Cited by
-
Structure and operation of the DNA-translocating type I DNA restriction enzymes.Genes Dev. 2012 Jan 1;26(1):92-104. doi: 10.1101/gad.179085.111. Genes Dev. 2012. PMID: 22215814 Free PMC article.
-
Low efficiency of homology-facilitated illegitimate recombination during conjugation in Escherichia coli.PLoS One. 2011;6(12):e28876. doi: 10.1371/journal.pone.0028876. Epub 2011 Dec 15. PLoS One. 2011. PMID: 22194937 Free PMC article.
-
The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein.Nucleic Acids Res. 2009 Feb;37(3):762-70. doi: 10.1093/nar/gkn988. Epub 2008 Dec 11. Nucleic Acids Res. 2009. PMID: 19074193 Free PMC article.
-
Highlights of the DNA cutters: a short history of the restriction enzymes.Nucleic Acids Res. 2014 Jan;42(1):3-19. doi: 10.1093/nar/gkt990. Epub 2013 Oct 18. Nucleic Acids Res. 2014. PMID: 24141096 Free PMC article.
-
DNA methylation by three Type I restriction modification systems of Escherichia coli does not influence gene regulation of the host bacterium.Nucleic Acids Res. 2021 Jul 21;49(13):7375-7388. doi: 10.1093/nar/gkab530. Nucleic Acids Res. 2021. PMID: 34181709 Free PMC article.
References
-
- Stent G.S. (1960) Papers on Bacterial Viruses. Little, Brown and Company, Boston, Toronto.
-
- Holmes F.L. (2001) Meselson, Stahl and the Replication of DNA. A History of the Most Beautiful Experiment in Biology. Yale University Press, New Haven and London.
-
- Murray N.E. (2002) Immigration control of DNA in bacteria: self versus non-self. Microbiology, 148, 3–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

