HMG-D and histone H1 alter the local accessibility of nucleosomal DNA
- PMID: 14654683
- PMCID: PMC291865
- DOI: 10.1093/nar/gkg923
HMG-D and histone H1 alter the local accessibility of nucleosomal DNA
Abstract
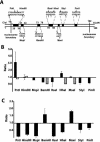
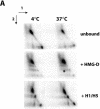
There is evidence that HMGB proteins facilitate, while linker histones inhibit chromatin remodelling, respectively. We have examined the effects of HMG-D and histone H1/H5 on accessibility of nucleosomal DNA. Using the 601.2 nucleosome positioning sequence designed by Widom and colleagues we assembled nucleosomes in vitro and probed DNA accessibility with restriction enzymes in the presence or absence of HMG-D and histone H1/H5. For HMG-D our results show increased digestion at two spatially adjacent sites, the dyad and one terminus of nucleosomal DNA. Elsewhere varying degrees of protection from digestion were observed. The C-terminal acidic tail of HMG-D is essential for this pattern of accessibility. Neither the HMG domain by itself nor in combination with the adjacent basic region is sufficient. Histone H1/H5 binding produces two sites of increased digestion on opposite faces of the nucleosome and decreased digestion at all other sites. Our results provide the first evidence of local changes in the accessibility of nucleosomal DNA upon separate interaction with two linker binding proteins.
Figures

Similar articles
-
A 'one-pot' assay for the accessibility of DNA in a nucleosome core particle.Nucleic Acids Res. 2004 Aug 25;32(15):e122. doi: 10.1093/nar/gnh121. Nucleic Acids Res. 2004. PMID: 15329384 Free PMC article.
-
The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits.J Mol Biol. 1994 Feb 11;236(1):189-98. doi: 10.1006/jmbi.1994.1128. J Mol Biol. 1994. PMID: 8107104
-
Interaction of maize chromatin-associated HMG proteins with mononucleosomes: role of core and linker histones.Biol Chem. 2003 Jul;384(7):1019-27. doi: 10.1515/BC.2003.114. Biol Chem. 2003. PMID: 12956418
-
Yeast HMO1: Linker Histone Reinvented.Microbiol Mol Biol Rev. 2016 Nov 30;81(1):e00037-16. doi: 10.1128/MMBR.00037-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2016. PMID: 27903656 Free PMC article. Review.
-
Linker histones versus HMG1/2: a struggle for dominance?Bioessays. 1998 Jul;20(7):584-8. doi: 10.1002/(SICI)1521-1878(199807)20:7<584::AID-BIES10>3.0.CO;2-W. Bioessays. 1998. PMID: 9723008 Review.
Cited by
-
Transient HMGB protein interactions with B-DNA duplexes and complexes.Biochem Biophys Res Commun. 2008 Jun 20;371(1):79-84. doi: 10.1016/j.bbrc.2008.04.024. Epub 2008 Apr 14. Biochem Biophys Res Commun. 2008. PMID: 18413230 Free PMC article.
-
HMGB1 deforms nucleosomal DNA to generate a dynamic chromatin environment counteracting the effects of linker histone.Sci Adv. 2025 Aug 15;11(33):eads4473. doi: 10.1126/sciadv.ads4473. Epub 2025 Aug 15. Sci Adv. 2025. PMID: 40815652 Free PMC article.
-
The Role of PARP1 and PAR in ATP-Independent Nucleosome Reorganisation during the DNA Damage Response.Genes (Basel). 2022 Dec 30;14(1):112. doi: 10.3390/genes14010112. Genes (Basel). 2022. PMID: 36672853 Free PMC article. Review.
-
High mobility group proteins HMGD and HMGZ interact genetically with the Brahma chromatin remodeling complex in Drosophila.Genetics. 2006 Feb;172(2):1069-78. doi: 10.1534/genetics.105.049957. Epub 2005 Nov 19. Genetics. 2006. PMID: 16299391 Free PMC article.
-
Characterization of the interaction between HMGB1 and H3-a possible means of positioning HMGB1 in chromatin.Nucleic Acids Res. 2014 Jan;42(2):848-59. doi: 10.1093/nar/gkt950. Epub 2013 Oct 23. Nucleic Acids Res. 2014. PMID: 24157840 Free PMC article.
References
-
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. - PubMed
-
- Anderson J.D. and Widom,J. (2000) Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol., 296, 979–987. - PubMed
-
- Polach K.J. and Widom,J. (1995) Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol., 254, 130–149. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

