Functional significance of intermediate cleavages in the 3'ETS of the pre-rRNA from Schizosaccharomyces pombe
- PMID: 14654686
- PMCID: PMC291872
- DOI: 10.1093/nar/gkg932
Functional significance of intermediate cleavages in the 3'ETS of the pre-rRNA from Schizosaccharomyces pombe
Abstract
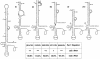
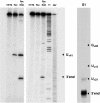
Pathways for the maturation of ribosomal RNAs are complex with numerous intermediate cleavage sites that are not always conserved closely in the course of evolution. Both in eukaryotes and bacteria genetic analyses and in vitro studies have strongly implicated RNase III-like enzymes in the processing of rRNA precursors. In Schizosacharomyces pombe, for example, the RNase III-like Pac1 nuclease has been shown to cleave the free 3'ETS at two known intermediate sites but, in the presence of RAC protein, the same RNA also is cleaved at the 3'-end of the 25 S rRNA sequence. In this study normal and mutant 3'ETS sequences were digested with the Pac1 enzyme to further evaluate its role in rRNA processing. Accurate cleavage at the known intermediate processing sites was dependent on the integrity of the helical structure at these sites as well as a more distal upper stem region in the conserved extended hairpin structure of the 3'ETS. The cleavage of mutant 3'ETS sequences also generally correlated with the known effects of these mutations on rRNA production, in vivo. One mutant, however, was efficiently processed in vivo but was not a substrate for the Pac1 nuclease, in vitro. In contrast, in the presence of RAC protein, the same RNA remained susceptible to Pac1 nuclease cleavage at the 3'-end of the 25 rRNA sequence, indicating that the removal of the 3'ETS does not require cleavage at the intermediate sites. These results suggest that basic maturation pathways may be less complex than previously reported raising similar questions about other intermediate processing sites, which have been identified by analyses of termini, and/or processing, in vitro.
Figures



References
-
- Ohtsuki K., Groner,Y. and Hurwitz,J. (1977) Isolation and purification of double-stranded ribonuclease from calf thymus. J. Biol. Chem., 252, 483–491. - PubMed
-
- Grummt I., Hall,S.H. and Crouch,R.J. (1979) Localization of an endonuclease specific for double-stranded RNA within the nucleolus and its implication in processing ribosomal transcripts. Eur. J. Biochem., 94, 437–443. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

