Effect of G-1 on histidine tRNA microhelix conformation
- PMID: 14654706
- PMCID: PMC291870
- DOI: 10.1093/nar/gkg930
Effect of G-1 on histidine tRNA microhelix conformation
Abstract
Histidine tRNAs (tRNA(His)) are unique in that they possess an extra 5'-base (G-1) not found in other tRNAs. Deletion of G-1 results in at least a 250-fold reduction in the rate of histidine charging in vitro. To better understand the role of the G-1 nucleotide in defining the structure of tRNA(His), and to correlate structure with cognate amino acid charging, NMR and molecular dynamics (MD) studies were performed on the wild-type and a DeltaG-1 mutant Escherichia coli histidine tRNA acceptor stem microhelix. Using NMR-derived distance restraints, global structural characteristics are described and interpreted to rationalize experimental observations with respect to aminoacylation activity. The quality of the NMR-derived solution conformations of the wild-type and DeltaG-1 histidine microhelices (micro helix(His)) is assessed using a variety of MD-based computational protocols. Most of the duplex regions of the acceptor stem and the UUCG tetraloop are well defined and effectively superimposable for the wild-type and DeltaG-1 mutant microhelix(His). Differences, however, are observed at the end of the helix and in the single-stranded CCCA-3' tail. The wild-type microhelix(His) structure is more well defined than the mutant and folds into a 'stacked fold-back' conformation. In contrast, we observe fraying of the first two base pairs and looping back of the single-stranded region in the DeltaG-1 mutant resulting in a much less well defined conformation. Thus the role of the extra G-1 base of the unique G-1:C73 base pair in tRNA(His) may be to prevent end-fraying and stabilize the stacked fold-back conformation of the CCCA-3' region.
Figures



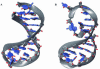
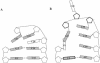
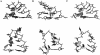
Similar articles
-
Enzymatic aminoacylation of an eight-base-pair microhelix with histidine.Proc Natl Acad Sci U S A. 1990 Nov;87(21):8655-9. doi: 10.1073/pnas.87.21.8655. Proc Natl Acad Sci U S A. 1990. PMID: 2236077 Free PMC article.
-
Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation.Nucleic Acids Res. 1989 Oct 11;17(19):7855-63. doi: 10.1093/nar/17.19.7855. Nucleic Acids Res. 1989. PMID: 2678006 Free PMC article.
-
RNA-binding site of Escherichia coli peptidyl-tRNA hydrolase.J Biol Chem. 2011 Nov 11;286(45):39585-94. doi: 10.1074/jbc.M111.281840. Epub 2011 Sep 19. J Biol Chem. 2011. PMID: 21930710 Free PMC article.
-
Striking effects of coupling mutations in the acceptor stem on recognition of tRNAs by Escherichia coli Met-tRNA synthetase and Met-tRNA transformylase.Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9262-6. doi: 10.1073/pnas.89.19.9262. Proc Natl Acad Sci U S A. 1992. PMID: 1409632 Free PMC article.
-
Functional compensation by particular nucleotide substitutions of a critical G*U wobble base-pair during aminoacylation of transfer RNA.J Mol Biol. 1999 Mar 5;286(4):1025-32. doi: 10.1006/jmbi.1999.2542. J Mol Biol. 1999. PMID: 10047479
Cited by
-
Human tRNA(Gly) acceptor-stem microhelix: crystallization and preliminary X-ray diffraction analysis at 1.2 A resolution.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Oct 1;63(Pt 10):858-61. doi: 10.1107/S1744309107041528. Epub 2007 Sep 19. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007. PMID: 17909289 Free PMC article.
-
The C nucleotide at the mature 5' end of the Escherichia coli proline tRNAs is required for the RNase E cleavage specificity at the 3' terminus as well as functionality.Nucleic Acids Res. 2022 Feb 22;50(3):1639-1649. doi: 10.1093/nar/gkab1260. Nucleic Acids Res. 2022. PMID: 35061897 Free PMC article.
-
The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase.RNA. 2010 May;16(5):1068-77. doi: 10.1261/rna.2087510. Epub 2010 Apr 1. RNA. 2010. PMID: 20360392 Free PMC article.
-
Crystallization and preliminary X-ray diffraction analysis of an Escherichia coli tRNA(Gly) acceptor-stem microhelix.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007 Jan 1;63(Pt 1):46-8. doi: 10.1107/S1744309106052870. Epub 2006 Dec 22. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007. PMID: 17183173 Free PMC article.
-
Molecular dynamics simulations of RNA: an in silico single molecule approach.Biopolymers. 2007 Feb 5;85(2):169-84. doi: 10.1002/bip.20620. Biopolymers. 2007. PMID: 17080418 Free PMC article. Review.
References
-
- Aberg A., Yaremchuk,A., Tukalo,M., Rasmussen,B. and Cusack,S. (1997) Crystal structure analysis of the activation of histidine by Thermus thermophilus histidyl-tRNA synthetase. Biochemistry, 36, 3084–3094. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

