Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome
- PMID: 14656962
- PMCID: PMC403796
- DOI: 10.1101/gr.1429003
Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome
Abstract
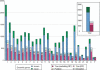
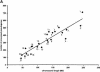
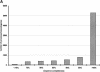
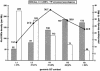
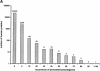
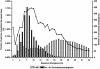
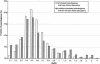
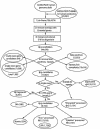
Processed pseudogenes were created by reverse-transcription of mRNAs; they provide snapshots of ancient genes existing millions of years ago in the genome. To find them in the present-day human, we developed a pipeline using features such as intron-absence, frame-disruption, polyadenylation, and truncation. This has enabled us to identify in recent genome drafts approximately 8000 processed pseudogenes (distributed from http://pseudogene.org). Overall, processed pseudogenes are very similar to their closest corresponding human gene, being 94% complete in coding regions, with sequence similarity of 75% for amino acids and 86% for nucleotides. Their chromosomal distribution appears random and dispersed, with the numbers on chromosomes proportional to length, suggesting sustained "bombardment" over evolution. However, it does vary with GC-content: Processed pseudogenes occur mostly in intermediate GC-content regions. This is similar to Alus but contrasts with functional genes and L1-repeats. Pseudogenes, moreover, have age profiles similar to Alus. The number of pseudogenes associated with a given gene follows a power-law relationship, with a few genes giving rise to many pseudogenes and most giving rise to few. The prevalence of processed pseudogenes agrees well with germ-line gene expression. Highly expressed ribosomal proteins account for approximately 20% of the total. Other notables include cyclophilin-A, keratin, GAPDH, and cytochrome c.
Figures





References
-
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J. 1994. Molecular biology of the cell. Garland Publishing, New York.
-
- Andersson, S.G., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133-140. - PubMed
-
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. - PubMed
WEB SITE REFERENCES
-
- http://bioinfo.mbb.yale.edu/genome/pseudogene; pseudogene database.
-
- http://www.ebi.ac.uk/GOA/; GO annotation of SWISS-PROT/TrEmbl proteins.
-
- http://www.ebi.ac.uk/proteome; EBI nonredundant human proteome.
-
- http://www.ebi.ac.uk/swissprot/; SWISS-PROT human protein sequences.
-
- http://www.ebi.ac.uk/trembl/; TrEMBL human protein sequences.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
